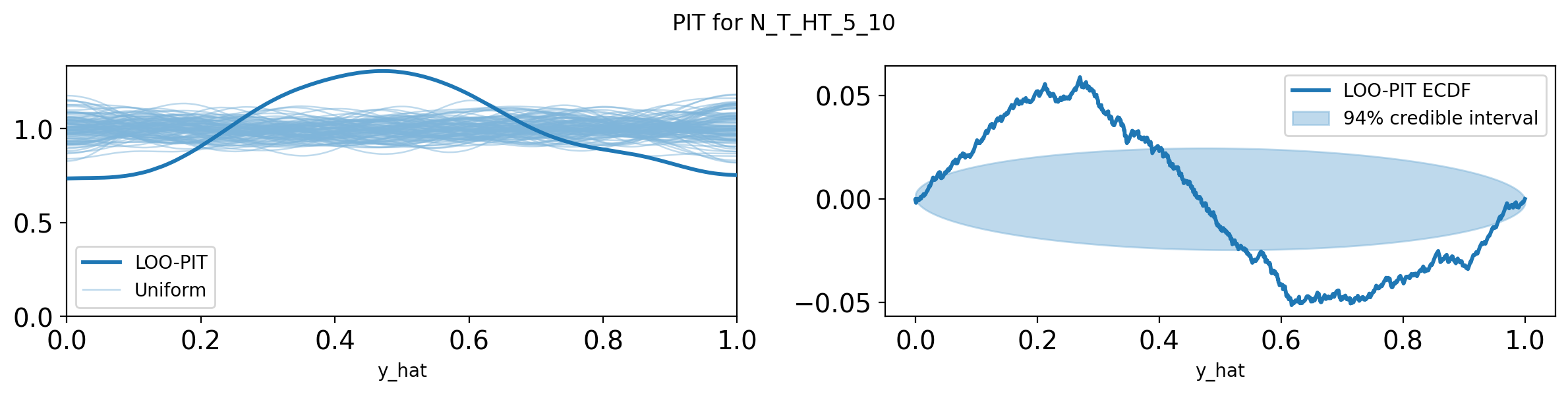
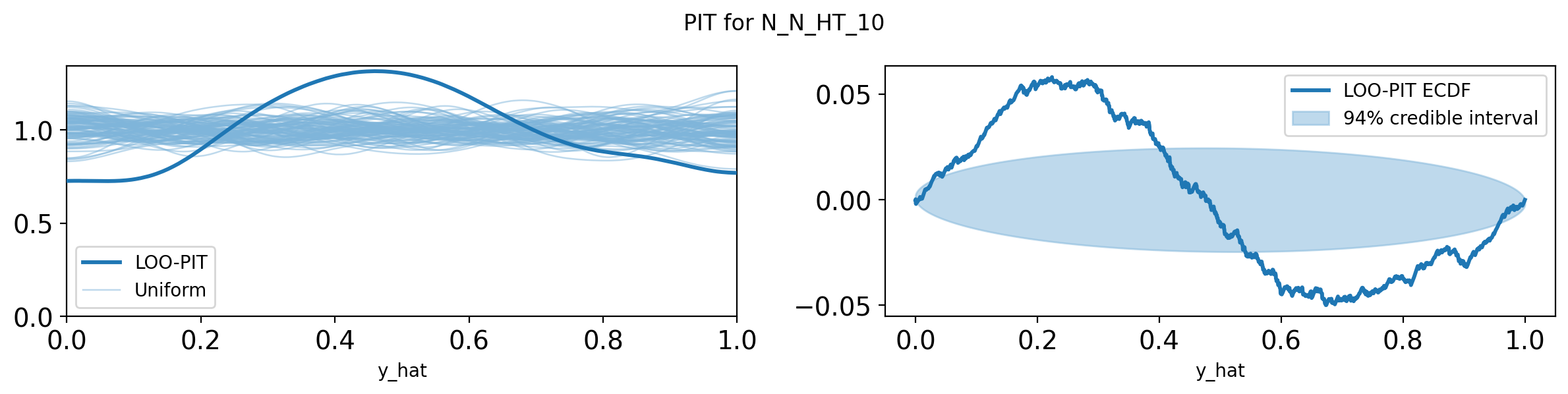
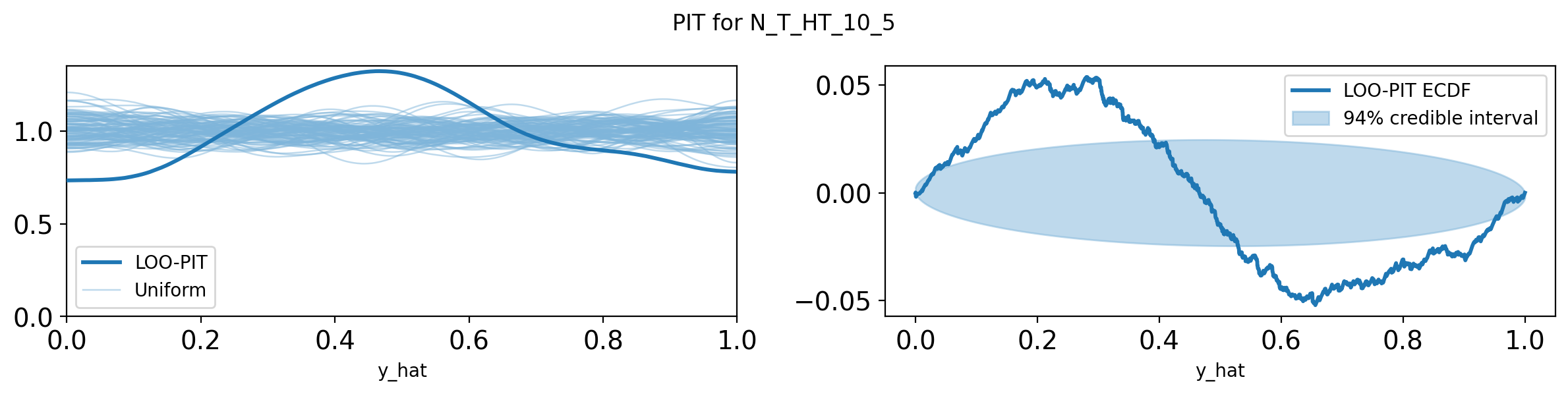
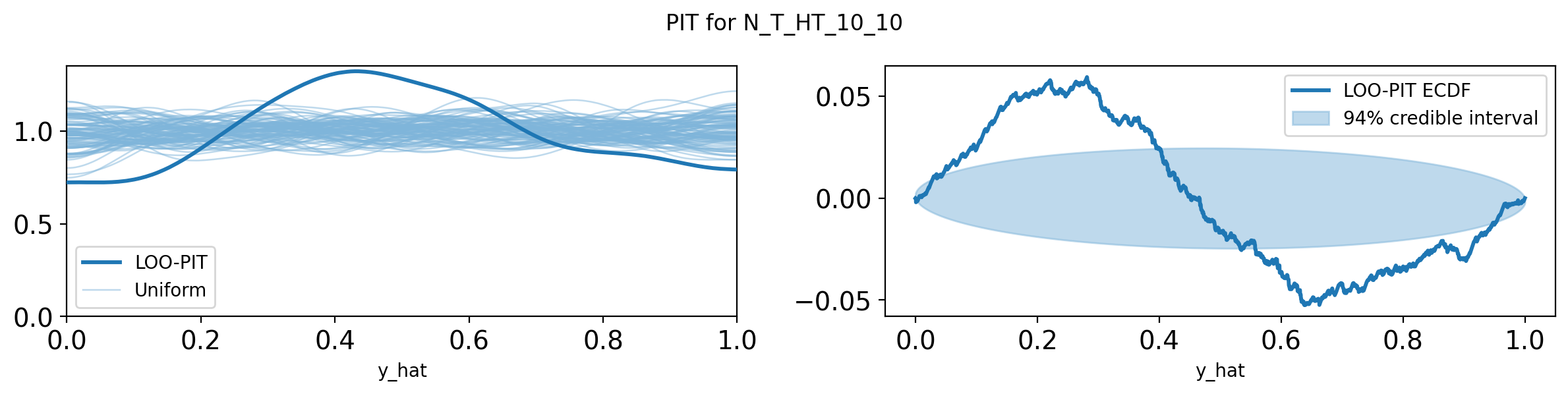
import pandas as pd
import numpy as np
import matplotlib.pyplot as plt
import matplotlib_inline
import pymc as pm
import arviz as az
import pytensor.tensor as pt
from scipy.stats import norm, t
from pprint import pprint
## Use a custom style for the plots
plt.style.use('smaller.mplstyle')
matplotlib_inline.backend_inline.set_matplotlib_formats('retina')
#%config InlineBackend.figure_formats = ['retina']Comparing GRW models for A/B Clustering
Quantifying differences in GRW models with varying parameters
Here, we compare probabilistic modeling of eigenvectors into A/B compartment calls for Hi-C data.
We compare different models, all using the same underlying structure, but varying smaller aspects of the model.
Goals
To quantify differences in model performance, we use the following metrics:
WAIC: Widely Applicable Information Criterion, a measure of model fit that penalizes complexity.
LOO: Leave-One-Out Cross-Validation, a method for estimating the predictive accuracy of a model.
We also try to assign uncertainty to the compartment calls, ultimately defining credible intervals for the compartment transitions.
Imports and setup
Load/show data
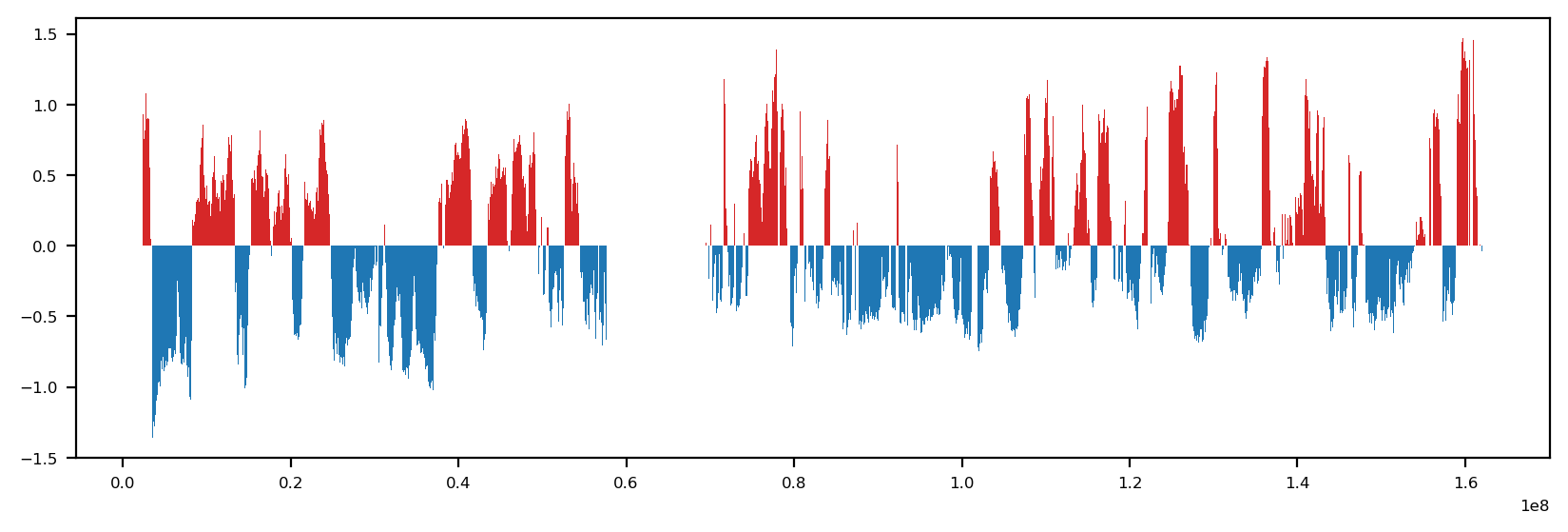
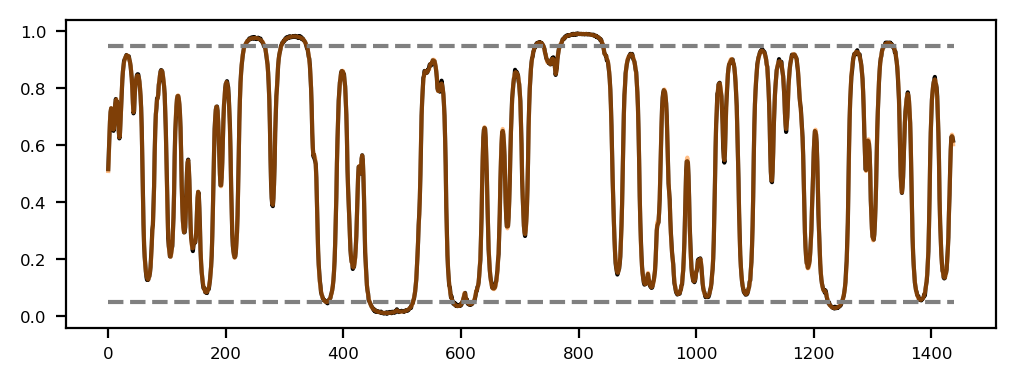
Here, we load the Hi-C contact matrix and the first eigenvector (E1) of the contact matrix. The E1 is used as a predictor for compartment assignment.
The eigendecomposition of the Hi-C matrix was performed with the workflow in the main folder (workflow.py), using the Open2C ecosystem
# Load the data
resolution = 100000
y = pd.Series(pd.read_csv(f"../data/eigs/sperm.eigs.{resolution}.cis.vecs.tsv", sep="\t")['E1'].values.flatten())
x = pd.Series(np.arange(0,y.shape[0])*resolution)
# Make a DataFrame object
df = pd.DataFrame({"start": x, "e1": y})
# Plot the data
fig, ax = plt.subplots(figsize=(10, 3))
ax.fill_between(df.start, df.e1, where=df.e1 > 0, color='tab:red', ec='None', label='E1', step='pre')
ax.fill_between(df.start, df.e1, where=df.e1 < 0, color='tab:blue', ec='None', label='E1', step='pre')
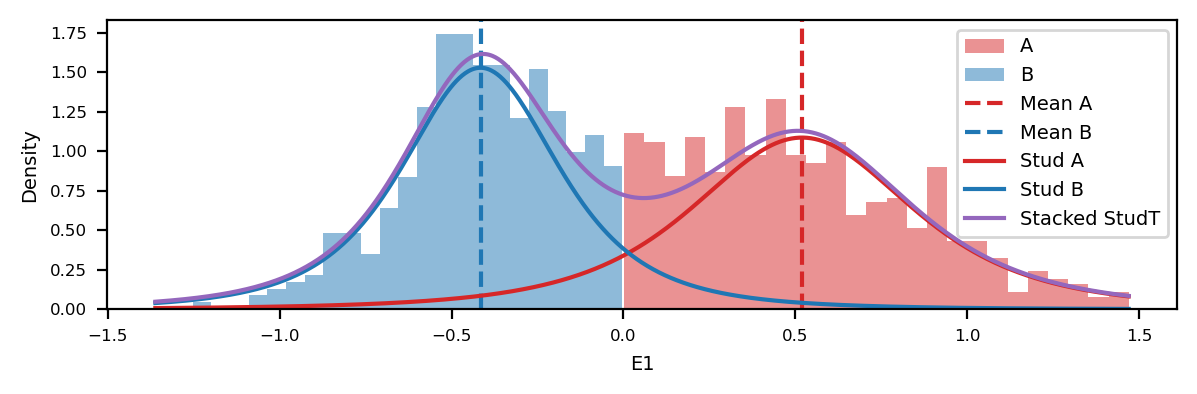
df.dropna(inplace=True)# Histogram of distribution of E1 values (A and B)
x = df["start"].values
y = df["e1"].values
dist_x_values = np.linspace(y.min(), y.max(), 1000)
a_dist = t.pdf(dist_x_values, loc=np.mean(y[y>0]), scale=y[y>0].std(), df=3)
b_dist = t.pdf(dist_x_values, loc=np.mean(y[y<0]), scale=y[y<0].std(), df=3)
f, ax = plt.subplots()
# A compartments
ax.hist(y[y>0], bins=25, color="tab:red", alpha=0.5, label="A", density=True)
# B compartments
ax.hist(y[y<0], bins=25, color="tab:blue", alpha=0.5, label="B", density=True)
# Mean values (vline)
ax.axvline(y[y>0].mean(), color="tab:red", linestyle="--", label="Mean A")
ax.axvline(y[y<0].mean(), color="tab:blue", linestyle="--", label="Mean B")
# Plot the normal distributions
ax.plot(dist_x_values, a_dist, color="tab:red", label="Stud A")
ax.plot(dist_x_values, b_dist, color="tab:blue", label="Stud B")
ax.plot(dist_x_values, a_dist+b_dist, color="tab:purple", label="Stacked StudT")
# Final touches
plt.xlabel("E1")
plt.ylabel("Density")
plt.legend(loc='best')
plt.tight_layout()Here, the observed distributions are compared to StudenT distributions of 3 degrees of freedom and the same mean and standard deviation as the observed data. The Student’s t-distribution is used because it is more robust with its heavier tails than the normal.
Models
Here, we expand on the results from 03_PyMC_GRW_logit.ipynb, and use the same model structure, but with different priors and likelihoods. Overall, the following thoughts are implemented:
- We assume that the eigenvectors can be drawn from a mixture of two distributions, one for each compartment (A and B). It could be any mixture, but the means should be negative for B and positive for A. We use either to model
mu:- Gaussians, as a simple starting point, or
- Student’s t-distributions, as a heavier-tailed alternative.
- The mixtures has to be ordered in PyMC to ensure that the A compartment is always positive and the B compartment is always negative.
- The mixture components have their own (learned) standard deviation,
sigma. - The mixture components use
muandsigma(each of shape 2) in either a,- Normal distribution, or
- Student’s t-distribution, with 10 degrees of freedom.
- We model the mixture weight as a non-centered (Gaussian) random walk prior, as a method of probabilistic smoothing of the PCA compartment calls.
epsis the (learned) step size of the random walk, and- either Normal or Student’s t-distributed
grw_sigmais the (learned) standard deviation of the random walk- either HalfNormal or HalfStudentT-distributed
- The logit-space (of the mixture weight) is then modeled deterministically as the cummulative sum of the random walk steps.
- The logit-space is then transformed to the probability space by squishing it through a sigmoid function.
- \(\hat{y}\) (
y_hat) is drawing the observations from the mixture distribution, using the mixture weight and the two components (the learned distributions of E1 values for A and B-compartments).
Model 1: T-dist in logit space, T-dist in mixture components
import pytensor.tensor as pt
with pm.Model(coords={"pos": df.index.values}) as latentT_Tmix_model:
e1 = pm.Data("e1", df.e1.values, dims="pos")
# Ordered parameters for the means of the two components
# Note: The ordered transform ensures that mu_a < mu_b
mu = pm.Normal("mu", mu=[-0.5, 0.5], sigma=0.3,
transform=pm.distributions.transforms.ordered, # IMPORTANT
shape=2,
)
sigma = pm.HalfNormal("sigma", 0.3, shape=2)
# GRW over logit space; non-centered reparameterization
grw_sigma = pm.HalfNormal("grw_sigma", 0.05)
# StudentT distribution for eps (got alot of divergences with nu=3)
eps = pm.StudentT("eps", nu=10, mu=0.0, sigma=1.0, dims="pos")
# Cumulative sum to create a Gaussian Random Walk
logit_w = pm.Deterministic("logit_w", pt.cumsum(eps * grw_sigma), dims="pos")
w = pm.Deterministic("w", pm.math.sigmoid(logit_w), dims="pos")
# Components of the mixture model
components = pm.StudentT.dist(nu=10, mu=mu, sigma=sigma, shape=2)
# Mixture model
# The observed data is modeled as a mixture of the two components
y_hat = pm.Mixture("y_hat", w=pm.math.stack([w,1-w], axis=1), comp_dists=components, observed=e1, dims='pos')Model 1: T-dist in logit space, Normal in mixture components
with pm.Model(coords={"pos": df.index.values}) as latentT_Nmix_model:
e1 = pm.Data("e1", df.e1.values, dims="pos")
# Ordered parameters for the means of the two components
# Note: The ordered transform ensures that mu_a < mu_b
mu = pm.Normal("mu", mu=[-0.5, 0.5], sigma=0.3,
transform=pm.distributions.transforms.ordered, # IMPORTANT
shape=2,
)
sigma = pm.HalfNormal("sigma", 0.3, shape=2)
# GRW over logit space; non-centered reparameterization
grw_sigma = pm.HalfNormal("grw_sigma", 0.05)
# StudentT distribution for eps (got alot of divergences with nu=3)
eps = pm.StudentT("eps", nu=10, mu=0.0, sigma=1.0, dims="pos")
# Cumulative sum to create a Gaussian Random Walk
logit_w = pm.Deterministic("logit_w", pt.cumsum(eps * grw_sigma), dims="pos")
w = pm.Deterministic("w", pm.math.sigmoid(logit_w), dims="pos")
# Components of the mixture model
components = pm.StudentT.dist(nu=10, mu=mu, sigma=sigma, shape=2)
# Mixture model
# The observed data is modeled as a mixture of the two components
y_hat = pm.Mixture("y_hat", w=pm.math.stack([w,1-w], axis=1), comp_dists=components, observed=e1, dims='pos')Parameter sweep configuration
Here is an attempt to sweep through parameters in a reproducible and deterministic way. The idea is to use a grid search over the parameters, and then run the models in parallel, and finally collect the results and analyze them.
First, let’s create a function that create a grid of parameters to sweep over using itertools.
Create parameter grid
import itertools as it
def create_model_grid():
"""
Create a grid of models for the different configurations as defined below.
Name the model according to the configuration for easier identification:
- comp_dist: Distribution of the components (Normal or StudentT)
- eps_dist: Distribution of the GRW steps (Normal or StudentT)
- grw_sigma_dist: Distribution of the GRW noise (HalfNormal or HalfStudentT)
name = f"{comp_dist}_{eps_dist}_{grw_sigma_dist}"
- If any distribution is StudentT, append the nu parameter to the name.
"""
def config_namer(comp_dist, comp_kwargs, eps_dist, eps_kwargs, grw_sigma_dist, grw_sigma_kwargs):
abbr = {
'Normal': 'N',
'StudentT': 'T',
'HalfNormal': 'HN',
'HalfStudentT': 'HT'
}
name = f"{abbr[comp_dist]}_{abbr[eps_dist]}_{abbr[grw_sigma_dist]}"
if comp_dist == 'StudentT':
name += f"_{comp_kwargs['nu']}"
if eps_dist == 'StudentT':
name += f"_{eps_kwargs['nu']}"
if grw_sigma_dist == 'HalfStudentT':
name += f"_{grw_sigma_kwargs['nu']}"
return name
comp_kwargs_list = [
('Normal', {}),
('StudentT', {'nu': 10}),
('StudentT', {'nu': 5})
]
eps_kwargs_list = [
('Normal', {'mu': 0.0, 'sigma': 1.0}),
('StudentT', {'nu': 10, 'mu': 0.0, 'sigma': 1.0}),
('StudentT', {'nu': 5, 'mu': 0.0, 'sigma': 0.5})
]
grw_sigma_kwargs_list = [
('HalfNormal', {'sigma': 0.05}),
('HalfStudentT', {'nu': 10, 'sigma': 0.05}),
('HalfStudentT', {'nu': 5, 'sigma': 0.01})
]
# Prior configurations for E1 mixture
mu_mu = [-0.5, 0.5]
mu_sigma = 0.3
sigma = 0.3
grid = it.product(comp_kwargs_list, eps_kwargs_list, grw_sigma_kwargs_list)
configs = []
for (comp_dist, comp_kwargs), (eps_dist, eps_kwargs), (grw_sigma_dist, grw_sigma_kwargs) in grid:
name = config_namer(
comp_dist, comp_kwargs,
eps_dist, eps_kwargs,
grw_sigma_dist, grw_sigma_kwargs
)
config = {
'name': name,
'comp_dist': comp_dist,
'comp_kwargs': comp_kwargs,
'eps_dist': eps_dist,
'eps_kwargs': eps_kwargs,
'grw_sigma_dist': grw_sigma_dist,
'grw_sigma_kwargs': grw_sigma_kwargs,
'mu_mu': mu_mu,
'mu_sigma': mu_sigma,
'sigma': sigma
}
configs.append(config)
return configs
from pprint import pprint
import json
# Instantiate the model grid
configs = create_model_grid()
pprint(configs)
# Save the model grid to a JSON file
with open("../results/model_grid.json", "w") as f:
json.dump(configs, f, indent=4)[{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_N_HN',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_N_HT_10',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_N_HT_5',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_T_HN_10',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_T_HT_10_10',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_T_HT_10_5',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_T_HN_5',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_T_HT_5_10',
'sigma': 0.3},
{'comp_dist': 'Normal',
'comp_kwargs': {},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'N_T_HT_5_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_N_HN_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_N_HT_10_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_N_HT_10_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HN_10_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_10_10_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_10_10_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HN_10_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_10_5_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 10},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_10_5_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_N_HN_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_N_HT_5_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'Normal',
'eps_kwargs': {'mu': 0.0, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_N_HT_5_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HN_5_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_5_10_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 10, 'sigma': 1.0},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_5_10_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfNormal',
'grw_sigma_kwargs': {'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HN_5_5',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 10, 'sigma': 0.05},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_5_5_10',
'sigma': 0.3},
{'comp_dist': 'StudentT',
'comp_kwargs': {'nu': 5},
'eps_dist': 'StudentT',
'eps_kwargs': {'mu': 0.0, 'nu': 5, 'sigma': 0.5},
'grw_sigma_dist': 'HalfStudentT',
'grw_sigma_kwargs': {'nu': 5, 'sigma': 0.01},
'mu_mu': [-0.5, 0.5],
'mu_sigma': 0.3,
'name': 'T_T_HT_5_5_5',
'sigma': 0.3}]Create model builder function
Then, we want to create a function (model_builder) that takes the parameters and returns a PyMC model. This function will be used to build the models in parallel.
NOTE: accidentally, I switched the order of mixture weights, so the posterior mean of
wnow determines the probability of being in a B compartment (coming from the distribution with negative mean)
def build_model_from_config(df, cfg):
"""
Build a PyMC model based on the provided configuration dictionary.
"""
with pm.Model(coords={"pos": df.index.values}) as model:
e1 = pm.Data("e1", df.e1.values, dims="pos")
mu = pm.Normal("mu", mu=cfg['mu_mu'], sigma=cfg['mu_sigma'],
transform=pm.distributions.transforms.ordered, shape=2)
sigma = pm.HalfNormal("sigma", sigma=cfg['sigma'], shape=2)
# components: Normal or StudentT
if cfg['comp_dist'] == "Normal":
components = pm.Normal.dist(mu=mu, sigma=sigma, shape=2)
else:
components = pm.StudentT.dist(mu=mu, sigma=sigma, **cfg['comp_kwargs'], shape=2)
# grw_sigma: HalfNormal or HalfStudentT
if cfg['grw_sigma_dist'] == "HalfNormal":
grw_sigma = pm.HalfNormal("grw_sigma", **cfg['grw_sigma_kwargs'])
else:
grw_sigma = pm.HalfStudentT("grw_sigma", **cfg['grw_sigma_kwargs'])
# eps: Normal or StudentT
if cfg['eps_dist'] == "Normal":
eps = pm.Normal("eps", dims="pos", **cfg['eps_kwargs'])
else:
eps = pm.StudentT("eps", dims="pos", **cfg['eps_kwargs'])
logit_w = pm.Deterministic("logit_w", pt.cumsum(eps * grw_sigma), dims="pos")
w = pm.Deterministic("w", pm.math.sigmoid(logit_w), dims="pos")
y_hat = pm.Mixture("y_hat", w=pt.stack([w, 1-w], axis=1), comp_dists=components,
observed=e1, dims='pos')
return modelSampling loop
Now, we create the sampling loop that should run the models in sequence.
We will name the models by joining the list [number, comp_dist, eps_dist, grw_sigma_dist][eps_nu, comp_nu].
All the traces (InferenceData) will be saved in a dictionary, model_traces.
Sample the models
from datetime import datetime
def log(msg, path=".04_PyMC_GRW_compare.log", overwrite=False):
print(f"LOG: {msg}")
if overwrite:
with open(path, "w") as f:
f.write(msg + "\n")
else:
with open(path, "a") as f:
f.write(msg + "\n")
# Reset the log file
log("Resetting log... \n", overwrite=True)
model_traces = {}
for i, cfg in enumerate(configs):
name = cfg['name']
if name in model_traces:
continue
else:
model_traces[name] = {'model': None, 'trace': None, 'scores': None}
log(f"Sampling {name}...")
try:
start = datetime.now()
log(f"Started at {start.strftime('%H:%M:%S')}")
model_traces[name]['model'] = build_model_from_config(df, cfg)
model = model_traces[name]['model']
trace = pm.sample(draws=1000, tune=1000, chains=7, cores=7, progressbar=False, model=model)
model_traces[name]['trace'] = trace
log(f"--> Took {(datetime.now()-start).total_seconds()} seconds!\n")
except Exception as e:
log(f"Model {name} failed: {e}\n")LOG: Resetting log...
LOG: Sampling N_N_HN...
LOG: Started at 22:36:58Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 302 seconds.LOG: --> Took 310.351568 seconds!
LOG: Sampling N_N_HT_10...
LOG: Started at 22:42:09Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 107 seconds.
There were 6980 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 118.167902 seconds!
LOG: Sampling N_N_HT_5...
LOG: Started at 22:44:07Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 134 seconds.
There were 6851 divergences after tuning. Increase `target_accept` or reparameterize.
Chain 6 reached the maximum tree depth. Increase `max_treedepth`, increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 143.521094 seconds!
LOG: Sampling N_T_HN_10...
LOG: Started at 22:46:30Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 320 seconds.LOG: --> Took 326.008741 seconds!
LOG: Sampling N_T_HT_10_10...
LOG: Started at 22:51:56Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 157 seconds.
There were 6843 divergences after tuning. Increase `target_accept` or reparameterize.
Chain 6 reached the maximum tree depth. Increase `max_treedepth`, increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 166.726683 seconds!
LOG: Sampling N_T_HT_10_5...
LOG: Started at 22:54:43Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 76 seconds.
There were 6997 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 85.394018 seconds!
LOG: Sampling N_T_HN_5...
LOG: Started at 22:56:08Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 229 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 240.948306 seconds!
LOG: Sampling N_T_HT_5_10...
LOG: Started at 23:00:09Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 164 seconds.
There were 6873 divergences after tuning. Increase `target_accept` or reparameterize.
Chain 4 reached the maximum tree depth. Increase `max_treedepth`, increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 174.36611 seconds!
LOG: Sampling N_T_HT_5_5...
LOG: Started at 23:03:04Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 91 seconds.
There were 6999 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 101.124336 seconds!
LOG: Sampling T_N_HN_10...
LOG: Started at 23:04:45Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 359 seconds.LOG: --> Took 365.130255 seconds!
LOG: Sampling T_N_HT_10_10...
LOG: Started at 23:10:50Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 122 seconds.
There were 6989 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 132.235343 seconds!
LOG: Sampling T_N_HT_10_5...
LOG: Started at 23:13:02Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 171 seconds.
There were 6805 divergences after tuning. Increase `target_accept` or reparameterize.
Chain 0 reached the maximum tree depth. Increase `max_treedepth`, increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 181.227185 seconds!
LOG: Sampling T_T_HN_10_10...
LOG: Started at 23:16:03Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 366 seconds.LOG: --> Took 372.410547 seconds!
LOG: Sampling T_T_HT_10_10_10...
LOG: Started at 23:22:16Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 103 seconds.
There were 6999 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 113.559086 seconds!
LOG: Sampling T_T_HT_10_10_5...
LOG: Started at 23:24:09Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 164 seconds.
There were 6969 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 173.920995 seconds!
LOG: Sampling T_T_HN_10_5...
LOG: Started at 23:27:03Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 241 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 246.952836 seconds!
LOG: Sampling T_T_HT_10_5_10...
LOG: Started at 23:31:10Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 120 seconds.
There were 6995 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 129.991988 seconds!
LOG: Sampling T_T_HT_10_5_5...
LOG: Started at 23:33:20Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 81 seconds.
There were 7000 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 91.027394 seconds!
LOG: Sampling T_N_HN_5...
LOG: Started at 23:34:51Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 362 seconds.LOG: --> Took 374.295864 seconds!
LOG: Sampling T_N_HT_5_10...
LOG: Started at 23:41:06Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 149 seconds.
There were 6938 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 158.387342 seconds!
LOG: Sampling T_N_HT_5_5...
LOG: Started at 23:43:44Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 95 seconds.
There were 6993 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 105.102596 seconds!
LOG: Sampling T_T_HN_5_10...
LOG: Started at 23:45:29Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 367 seconds.LOG: --> Took 372.963417 seconds!
LOG: Sampling T_T_HT_5_10_10...
LOG: Started at 23:51:42Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 110 seconds.
There were 6978 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 120.264809 seconds!
LOG: Sampling T_T_HT_5_10_5...
LOG: Started at 23:53:42Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 92 seconds.
There were 7000 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 102.415237 seconds!
LOG: Sampling T_T_HN_5_5...
LOG: Started at 23:55:25Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 229 seconds.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 235.065423 seconds!
LOG: Sampling T_T_HT_5_5_10...
LOG: Started at 23:59:20Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 115 seconds.
There were 6980 divergences after tuning. Increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 125.419846 seconds!
LOG: Sampling T_T_HT_5_5_5...
LOG: Started at 00:01:25Initializing NUTS using jitter+adapt_diag...
Multiprocess sampling (7 chains in 7 jobs)
NUTS: [mu, sigma, grw_sigma, eps]
Sampling 7 chains for 1_000 tune and 1_000 draw iterations (7_000 + 7_000 draws total) took 148 seconds.
There were 6826 divergences after tuning. Increase `target_accept` or reparameterize.
Chain 4 reached the maximum tree depth. Increase `max_treedepth`, increase `target_accept` or reparameterize.
The rhat statistic is larger than 1.01 for some parameters. This indicates problems during sampling. See https://arxiv.org/abs/1903.08008 for details
The effective sample size per chain is smaller than 100 for some parameters. A higher number is needed for reliable rhat and ess computation. See https://arxiv.org/abs/1903.08008 for detailsLOG: --> Took 158.14628 seconds!
model_traces.items()dict_items([('N_N_HN', {'model': <pymc.model.core.Model object at 0x149177a5b4d0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_N_HT_10', {'model': <pymc.model.core.Model object at 0x14919e1710f0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_N_HT_5', {'model': <pymc.model.core.Model object at 0x1491912d3490>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_T_HN_10', {'model': <pymc.model.core.Model object at 0x1491912d2190>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_T_HT_10_10', {'model': <pymc.model.core.Model object at 0x1492013468b0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_T_HT_10_5', {'model': <pymc.model.core.Model object at 0x14917608fce0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_T_HN_5', {'model': <pymc.model.core.Model object at 0x14917608df30>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_T_HT_5_10', {'model': <pymc.model.core.Model object at 0x14917608de00>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('N_T_HT_5_5', {'model': <pymc.model.core.Model object at 0x14917608d6e0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_N_HN_10', {'model': <pymc.model.core.Model object at 0x14917608e520>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_N_HT_10_10', {'model': <pymc.model.core.Model object at 0x14917608e2c0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_N_HT_10_5', {'model': <pymc.model.core.Model object at 0x14917608e3f0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HN_10_10', {'model': <pymc.model.core.Model object at 0x14917608d940>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_10_10_10', {'model': <pymc.model.core.Model object at 0x14917608e780>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_10_10_5', {'model': <pymc.model.core.Model object at 0x14917608fbb0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HN_10_5', {'model': <pymc.model.core.Model object at 0x14917608dba0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_10_5_10', {'model': <pymc.model.core.Model object at 0x14917608e190>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_10_5_5', {'model': <pymc.model.core.Model object at 0x14917608f230>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_N_HN_5', {'model': <pymc.model.core.Model object at 0x14917608d810>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_N_HT_5_10', {'model': <pymc.model.core.Model object at 0x14917608dcd0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_N_HT_5_5', {'model': <pymc.model.core.Model object at 0x1491676a48a0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HN_5_10', {'model': <pymc.model.core.Model object at 0x14917608f5c0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_5_10_10', {'model': <pymc.model.core.Model object at 0x14917608efd0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_5_10_5', {'model': <pymc.model.core.Model object at 0x14917608fa80>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HN_5_5', {'model': <pymc.model.core.Model object at 0x14917608e650>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_5_5_10', {'model': <pymc.model.core.Model object at 0x1491676a56e0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None}), ('T_T_HT_5_5_5', {'model': <pymc.model.core.Model object at 0x1491676a5ba0>, 'trace': Inference data with groups:
> posterior
> sample_stats
> observed_data, 'scores': None})])Log-lik, WAIC and PPC
for name, model_dict in model_traces.items():
# model_dict.keys(): {'model', 'trace', 'scores'}
try:
print(name)
pm.compute_log_likelihood(
model_dict['trace'],
model=model_dict['model'],
progressbar=False)
except Exception as e:
print(f"{name} failed computing log_likelihood: {e}")
try:
prior_predictive = pm.sample_prior_predictive(
1000,
model=model_dict['model'])
model_dict['trace'].extend(prior_predictive)
except Exception as e:
print(f"{name} failed sampling prior predictive: {e}")
try:
posterior_predictive = pm.sample_posterior_predictive(
model_dict['trace'],
model=model_dict['model'],
progressbar=False)
model_dict['trace'].extend(posterior_predictive)
except Exception as e:
print(f"{name} failed sampling posterior predictive: {e}")
try:
loo = az.loo(model_dict['trace'])
waic = az.waic(model_dict['trace'])
model_dict['scores'] = {
"loo": loo,
"waic": waic
}
except Exception as e:
print(f"{name} failed during loo/waic: {e}")N_N_HNSampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_N_HT_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_N_HT_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_T_HN_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_T_HT_10_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_T_HT_10_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_T_HN_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_T_HT_5_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(N_T_HT_5_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_N_HN_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_N_HT_10_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_N_HT_10_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HN_10_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_10_10_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_10_10_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HN_10_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_10_5_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_10_5_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_N_HN_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_N_HT_5_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_N_HT_5_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HN_5_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_5_10_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_5_10_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HN_5_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]T_T_HT_5_5_10Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(T_T_HT_5_5_5Sampling: [eps, grw_sigma, mu, sigma, y_hat]
Sampling: [y_hat]
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:797: UserWarning: Estimated shape parameter of Pareto distribution is greater than 0.70 for one or more samples. You should consider using a more robust model, this is because importance sampling is less likely to work well if the marginal posterior and LOO posterior are very different. This is more likely to happen with a non-robust model and highly influential observations.
warnings.warn(
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1655: UserWarning: For one or more samples the posterior variance of the log predictive densities exceeds 0.4. This could be indication of WAIC starting to fail.
See http://arxiv.org/abs/1507.04544 for details
warnings.warn(Save the results
import os.path as op
import os
import cloudpickle
# Save the traces to file (netcdf)
dir = "../results"
for name, d in model_traces.items():
# Save the model.Model as a cloudpickle.dumps
os.makedirs(op.join(dir, "models"), exist_ok=True)
model_path = op.join(dir, "models", f"{name}_model.pkl")
# print(model_path)
with open(model_path, "wb") as f:
cloudpickle.dump(d['model'], f)
# Save the trace as a netcdf file
trace_path = op.join(dir, "traces", f"{name}_trace.nc")
os.makedirs(op.dirname(trace_path), exist_ok=True)
az.to_netcdf(d['trace'], trace_path)
# Load model
with open("../results/models/N_N_HN_model.pkl", "rb") as f:
test_model = cloudpickle.load(f)
# Load trace
test_trace = az.from_netcdf("../results/traces/N_N_HN_trace.nc")from math import ceil
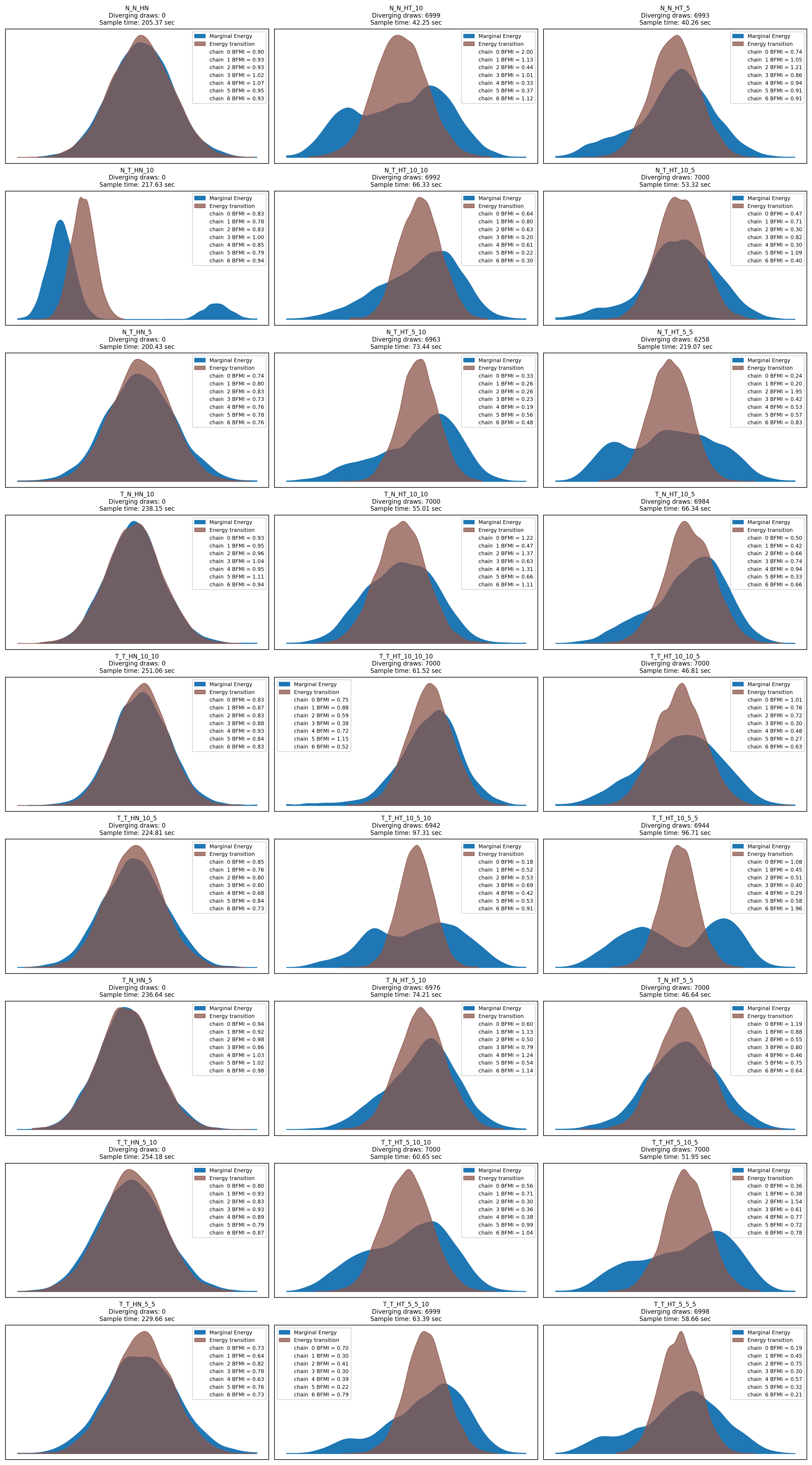
# Plot the energy for all traces
ncol = 3
nrows = ceil(len(model_traces) / ncol)
f, axs = plt.subplots(nrows=nrows, ncols=ncol, figsize=(15, 3*nrows))
axs = axs.flatten()
for i, (name, d) in enumerate(model_traces.items()):
ax = axs[i]
az.plot_energy(d['trace'], ax=ax)
ax.set_title(name + "\n" +
f"Diverging draws: {d['trace'].sample_stats.diverging.sum().values}\n" +
f"Sample time: {d['trace'].sample_stats.sampling_time:.2f} sec"
)
# Remove empty subplots
[f.delaxes(ax) for ax in axs if not ax.has_data()]
plt.tight_layout()
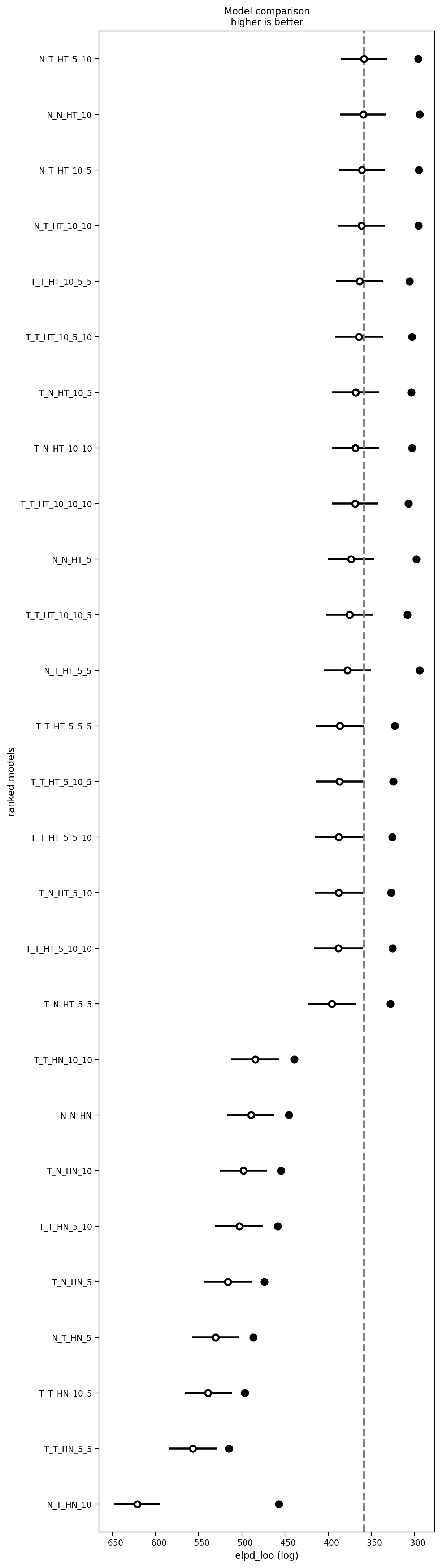
# az.plot_energy(test_trace);# Combine into InferenceData comparison object
loo_dict = {name: model_traces[name]['scores']['loo'] for name in model_traces.keys()}
cmp_df = az.compare(loo_dict, method="stacking", ic="loo")
# Save the comparison DataFrame
cmp_df.to_csv("../results/model_comparison.csv", )
# Print the comparison DataFrame
print("\nModel Comparison Results:")
cmp_df
Model Comparison Results:| rank | elpd_loo | p_loo | elpd_diff | weight | se | dse | warning | scale | |
|---|---|---|---|---|---|---|---|---|---|
| N_T_HT_5_10 | 0 | -13.942824 | 112.407689 | 0.000000 | 0.501709 | 26.172102 | 0.000000 | True | log |
| N_T_HT_10_10 | 1 | -14.232323 | 115.634082 | 0.289498 | 0.232978 | 25.852563 | 4.944408 | True | log |
| N_T_HT_10_5 | 2 | -15.196429 | 113.624439 | 1.253604 | 0.265313 | 25.562472 | 5.420416 | True | log |
| T_N_HT_10_5 | 3 | -29.299767 | 103.590498 | 15.356943 | 0.000000 | 26.574965 | 5.712471 | True | log |
| T_T_HT_10_10_10 | 4 | -30.656224 | 110.373495 | 16.713400 | 0.000000 | 26.435805 | 6.042163 | True | log |
| N_T_HT_5_5 | 5 | -33.954243 | 134.999306 | 20.011419 | 0.000000 | 26.360560 | 7.147779 | True | log |
| T_N_HT_10_10 | 6 | -39.808868 | 110.980355 | 25.866044 | 0.000000 | 26.492594 | 5.556646 | True | log |
| T_T_HT_10_5_5 | 7 | -46.613526 | 115.610006 | 32.670702 | 0.000000 | 26.806032 | 6.565634 | True | log |
| T_T_HT_10_10_5 | 8 | -48.198680 | 120.920450 | 34.255855 | 0.000000 | 26.793137 | 7.001829 | True | log |
| T_T_HT_5_10_5 | 9 | -59.106328 | 105.086513 | 45.163503 | 0.000000 | 27.142046 | 6.035451 | True | log |
| T_T_HT_5_5_5 | 10 | -59.685208 | 102.204978 | 45.742384 | 0.000000 | 27.823967 | 7.678840 | True | log |
| T_T_HT_5_10_10 | 11 | -60.728334 | 102.599512 | 46.785510 | 0.000000 | 27.133034 | 6.249436 | True | log |
| T_N_HT_5_5 | 12 | -67.525844 | 107.832978 | 53.583020 | 0.000000 | 27.624499 | 7.079179 | True | log |
| N_T_HN_10 | 13 | -173.060588 | 68.185244 | 159.117764 | 0.000000 | 25.601244 | 7.208652 | False | log |
| N_N_HT_10 | 14 | -174.314027 | 221.807800 | 160.371203 | 0.000000 | 25.234972 | 7.764120 | True | log |
| N_N_HT_5 | 15 | -183.304207 | 230.436980 | 169.361383 | 0.000000 | 25.714858 | 8.332182 | True | log |
| N_N_HN | 16 | -190.088696 | 65.218096 | 176.145872 | 0.000000 | 25.583171 | 7.596365 | False | log |
| T_T_HN_10_10 | 17 | -192.626419 | 65.398645 | 178.683595 | 0.000000 | 26.146340 | 7.334733 | False | log |
| T_T_HT_10_5_10 | 18 | -196.046002 | 215.026323 | 182.103178 | 0.000000 | 26.522684 | 6.822517 | True | log |
| T_N_HN_10 | 19 | -208.959314 | 61.984608 | 195.016489 | 0.000000 | 26.085043 | 7.693501 | False | log |
| T_T_HN_5_10 | 20 | -216.997040 | 62.653391 | 203.054216 | 0.000000 | 26.711356 | 7.740838 | False | log |
| T_N_HT_5_10 | 21 | -229.496264 | 218.853051 | 215.553440 | 0.000000 | 27.049425 | 7.853962 | True | log |
| T_T_HT_5_5_10 | 22 | -230.189140 | 214.764648 | 216.246316 | 0.000000 | 27.201740 | 7.730539 | True | log |
| T_N_HN_5 | 23 | -233.133380 | 59.696692 | 219.190556 | 0.000000 | 26.665638 | 8.054481 | False | log |
| N_T_HN_5 | 24 | -248.998777 | 58.803980 | 235.055953 | 0.000000 | 25.585742 | 9.124896 | False | log |
| T_T_HN_10_5 | 25 | -267.526199 | 56.568757 | 253.583375 | 0.000000 | 26.100456 | 9.161338 | True | log |
| T_T_HN_5_5 | 26 | -291.694932 | 54.513089 | 277.752108 | 0.000000 | 26.616132 | 9.474451 | False | log |
Import and analyze the results
import pandas as pd
import arviz as az
from pathlib import Path
from cloudpickle import load
from glob import glob
# Load models
model_traces = {}
for pkl in glob("../results/models/*.pkl"):
name = Path(pkl).name.removesuffix("_model.pkl")
trace_path = f"../results/traces/{name}_trace.nc"
model_traces[name] = {}
with open(pkl, "rb") as f:
model_traces[name]['model'] = load(f)
model_traces[name]['trace'] = az.from_netcdf(trace_path)
# Load the comparison DataFrame
cmp_df = pd.read_csv("../results/model_comparison.csv", index_col=0)
import matplotlib.pyplot as plt
top_n = 8
for name in cmp_df.index[:top_n]:
f, axs = plt.subplots(1,2, figsize=(12, 3))
trace = model_traces[name]['trace']
az.plot_loo_pit(trace, y='y_hat', ax=axs[0])
az.plot_loo_pit(trace, y='y_hat', ecdf=True, ax=axs[1])
f.suptitle(f"PIT for {name}")
f.tight_layout()/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/arviz/stats/stats.py:1045: RuntimeWarning: overflow encountered in exp
weights = 1 / np.exp(len_scale - len_scale[:, None]).sum(axis=1)
/home/sojern/miniconda3/envs/pymc/lib/python3.13/site-packages/numpy/_core/_methods.py:52: RuntimeWarning: overflow encountered in reduce
return umr_sum(a, axis, dtype, out, keepdims, initial, where)Test: Post-hoc decoding of mixture weight
import arviz as az
import cloudpickle as cp
name = "N_N_HN"
trace = az.from_netcdf(f"../results/traces/{name}_trace.nc")
with open(f"../results/models/{name}_model.pkl", "rb") as f:
model = cp.load(f)trace-
<xarray.Dataset> Size: 242MB Dimensions: (chain: 7, draw: 1000, pos: 1438, mu_dim_0: 2, sigma_dim_0: 2) Coordinates: * chain (chain) int64 56B 0 1 2 3 4 5 6 * draw (draw) int64 8kB 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 * pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621 * mu_dim_0 (mu_dim_0) int64 16B 0 1 * sigma_dim_0 (sigma_dim_0) int64 16B 0 1 Data variables: eps (chain, draw, pos) float64 81MB ... mu (chain, draw, mu_dim_0) float64 112kB ... sigma (chain, draw, sigma_dim_0) float64 112kB ... grw_sigma (chain, draw) float64 56kB ... logit_w (chain, draw, pos) float64 81MB ... w (chain, draw, pos) float64 81MB ... Attributes: created_at: 2025-07-17T20:42:05.061371+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 sampling_time: 302.33364176750183 tuning_steps: 1000 -
<xarray.Dataset> Size: 81MB Dimensions: (chain: 7, draw: 1000, pos: 1438) Coordinates: * chain (chain) int64 56B 0 1 2 3 4 5 6 * draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621 Data variables: y_hat (chain, draw, pos) float64 81MB ... Attributes: created_at: 2025-07-17T22:04:45.542489+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 -
<xarray.Dataset> Size: 81MB Dimensions: (chain: 7, draw: 1000, pos: 1438) Coordinates: * chain (chain) int64 56B 0 1 2 3 4 5 6 * draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621 Data variables: y_hat (chain, draw, pos) float64 81MB ... Attributes: created_at: 2025-07-17T22:04:05.221796+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 -
<xarray.Dataset> Size: 862kB Dimensions: (chain: 7, draw: 1000) Coordinates: * chain (chain) int64 56B 0 1 2 3 4 5 6 * draw (draw) int64 8kB 0 1 2 3 4 5 ... 995 996 997 998 999 Data variables: (12/17) tree_depth (chain, draw) int64 56kB ... largest_eigval (chain, draw) float64 56kB ... step_size_bar (chain, draw) float64 56kB ... process_time_diff (chain, draw) float64 56kB ... diverging (chain, draw) bool 7kB ... step_size (chain, draw) float64 56kB ... ... ... acceptance_rate (chain, draw) float64 56kB ... index_in_trajectory (chain, draw) int64 56kB ... perf_counter_start (chain, draw) float64 56kB ... max_energy_error (chain, draw) float64 56kB ... lp (chain, draw) float64 56kB ... perf_counter_diff (chain, draw) float64 56kB ... Attributes: created_at: 2025-07-17T20:42:05.081859+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 sampling_time: 302.33364176750183 tuning_steps: 1000 -
<xarray.Dataset> Size: 35MB Dimensions: (chain: 1, draw: 1000, pos: 1438, sigma_dim_0: 2, mu_dim_0: 2) Coordinates: * chain (chain) int64 8B 0 * draw (draw) int64 8kB 0 1 2 3 4 5 6 ... 993 994 995 996 997 998 999 * pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621 * sigma_dim_0 (sigma_dim_0) int64 16B 0 1 * mu_dim_0 (mu_dim_0) int64 16B 0 1 Data variables: grw_sigma (chain, draw) float64 8kB ... w (chain, draw, pos) float64 12MB ... sigma (chain, draw, sigma_dim_0) float64 16kB ... eps (chain, draw, pos) float64 12MB ... logit_w (chain, draw, pos) float64 12MB ... mu (chain, draw, mu_dim_0) float64 16kB ... Attributes: created_at: 2025-07-17T22:04:10.448419+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 -
<xarray.Dataset> Size: 12MB Dimensions: (chain: 1, draw: 1000, pos: 1438) Coordinates: * chain (chain) int64 8B 0 * draw (draw) int64 8kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999 * pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621 Data variables: y_hat (chain, draw, pos) float64 12MB ... Attributes: created_at: 2025-07-17T22:04:10.455784+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0 -
<xarray.Dataset> Size: 23kB Dimensions: (pos: 1438) Coordinates: * pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621 Data variables: y_hat (pos) float64 12kB ... Attributes: created_at: 2025-07-17T20:42:05.086062+00:00 arviz_version: 0.21.0 inference_library: pymc inference_library_version: 5.22.0
model\[ \begin{array}{rcl} \text{mu} &\sim & \operatorname{Normal}(\text{<constant>},~0.3)\\\text{sigma} &\sim & \operatorname{HalfNormal}(0,~0.3)\\\text{grw\_sigma} &\sim & \operatorname{HalfNormal}(0,~0.05)\\\text{eps} &\sim & \operatorname{Normal}(0,~1)\\\text{logit\_w} &\sim & \operatorname{Deterministic}(f(\text{eps},~\text{grw\_sigma}))\\\text{w} &\sim & \operatorname{Deterministic}(f(\text{eps},~\text{grw\_sigma}))\\\text{y\_hat} &\sim & \operatorname{MarginalMixture}(f(\text{eps},~\text{grw\_sigma}),~\operatorname{Normal}(\text{mu},~\text{sigma}))\\\text{y\_hat} &\sim & \operatorname{MarginalMixture}(f(\text{eps},~\text{grw\_sigma}),~\operatorname{Normal}(\text{mu},~\text{sigma})) \end{array} \]
# Just check what the xarray dims look like
trace.posterior['w'].stack(dims=['chain', 'draw'])<xarray.DataArray 'w' (pos: 1438, dims: 7000)> Size: 81MB
array([[0.44937833, 0.58546809, 0.43819027, ..., 0.51989126, 0.41636437,
0.43202614],
[0.52977449, 0.62375669, 0.58263025, ..., 0.50868668, 0.50093716,
0.5130883 ],
[0.5916006 , 0.76048527, 0.67466505, ..., 0.70195694, 0.43678679,
0.44905003],
...,
[0.80690199, 0.73596005, 0.43482456, ..., 0.70743037, 0.26833616,
0.3913885 ],
[0.8867993 , 0.56482857, 0.4902178 , ..., 0.77131705, 0.24841057,
0.35699746],
[0.82271155, 0.61978168, 0.47971009, ..., 0.77320498, 0.2004193 ,
0.30180627]], shape=(1438, 7000))
Coordinates:
* pos (pos) int64 12kB 24 25 26 27 28 29 ... 1615 1618 1619 1620 1621
* dims (dims) object 56kB MultiIndex
* chain (dims) int64 56kB 0 0 0 0 0 0 0 0 0 0 0 0 ... 6 6 6 6 6 6 6 6 6 6 6
* draw (dims) int64 56kB 0 1 2 3 4 5 6 7 ... 993 994 995 996 997 998 999Now, we can flatten the weights to a n_chains*ndraws X n_bins array and assign an underlying distribution with a Bernoulli trial on each draw.
Note, that the mean of the Bernoulli samples from the posterior draws will converge towards the posterior mean of
w. It is thus only an unnecessary step that adds noise and not addition information.
import numpy as np
# Extract posterior samples of w
w_flat = trace.posterior['w'].stack(dims=['chain', 'draw'])
w_mean = trace.posterior['w'].mean(dim=['chain', 'draw'])
# Sample z draws
seed = 42
rng = np.random.default_rng(seed)
z_samples = rng.binomial(1, w_flat) # shape (samples, pos)
# Compute posterior probability of z=1
z_probs = z_samples.mean(axis=1) # per-position probabilityz_samplesarray([[1, 1, 1, ..., 1, 1, 1],
[0, 0, 0, ..., 1, 1, 1],
[1, 1, 0, ..., 0, 1, 0],
...,
[1, 1, 1, ..., 1, 0, 0],
[1, 0, 0, ..., 0, 0, 0],
[1, 1, 0, ..., 1, 1, 0]], shape=(1438, 7000))z_probs
# Plots as a quick curve
plt.plot(z_probs, color='black', label='Posterior Probability of z=1')
plt.plot(w_mean, color='tab:orange', label='Posterior Mean of w', alpha = 0.5)
plt.hlines([0.95,0.05], xmin=0, xmax=len(z_probs), colors='gray', linestyles='--', label='95% CI');Try some other logic:
# Logic: We define the threshold for a certain assignment of either
# A: z_posterior > A_threshold, or B: z_posterior < B_threshold
# The rest will be considered in-transition and could be visualized as a gradient or with shaded regions
A_threshold = 0.05 # A = 0 in the binary assignment
B_threshold = 0.95 # B = 1 in the binary assignment
# Assign compartments based on the threshold
z_assignment = np.where(z_probs < A_threshold, 0,
np.where(z_probs > B_threshold, 1, np.nan))
# Turn around the assignment to have 1 for A and -1 for B
z_assignment = np.where(z_assignment == 0, 1,
np.where(z_assignment == 1, -1, np.nan))
# Plot as quick curve
plt.plot(z_assignment, color='tab:purple', label='Posterior Probability of z=1')
plt.hlines([0.95,0.05], xmin=0, xmax=len(z_assignment), colors='gray', linestyles='--', label='95% CI');
import pandas as pd
import matplotlib.pyplot as plt
import matplotlib_inline
from matplotlib.collections import PatchCollection
from matplotlib.patches import Rectangle
matplotlib_inline.backend_inline.set_matplotlib_formats('svg')
# Load the data
resolution = 100000
y = pd.Series(pd.read_csv(f"../data/eigs/fibroblast.eigs.{resolution}.cis.vecs.tsv", sep="\t")['E1'].values.flatten())
x = pd.Series(np.arange(0,y.shape[0])*resolution)
# Make a DataFrame object
df = pd.DataFrame({"start": x, "e1": y})
df.dropna(inplace=True)
# Quick Plot:
fig, ax = plt.subplots(figsize=(10, 3))
ax.fill_between(df.start, df.e1, where=df.e1 > 0, color='tab:red', ec='None', label='A', step='pre')
ax.fill_between(df.start, df.e1, where=df.e1 < 0, color='tab:blue', ec='None', label='B', step='pre')
ax.plot(df.start, z_assignment, color='tab:green', lw=1, label='z assignment')
# Add shaded areas for the nans
patches = []
in_nan = False
start = None
for i in range(len(z_assignment)):
if np.isnan(z_assignment[i]) and not in_nan:
# Start of NaN block
in_nan = True
start = df.start.iloc[i]
elif not np.isnan(z_assignment[i]) and in_nan:
# End of NaN block
end = df.start.iloc[i]
patches.append(Rectangle((start, -1), end - start, 2, alpha=0.2, color='gray'))
in_nan = False
# If ending on NaNs
if in_nan:
end = df.start.iloc[-1] + resolution
patches.append(Rectangle((start, -1), end - start, 2, alpha=1, color='gray'))
ax.add_collection(PatchCollection(patches, match_original=True))
ax.set_xlabel("Genomic Position")
ax.set_ylabel("E1")
ax.legend(loc='lower right')
plt.tight_layout()
--------------------------------------------------------------------------- ValueError Traceback (most recent call last) Cell In[26], line 24 22 ax.fill_between(df.start, df.e1, where=df.e1 > 0, color='tab:red', ec='None', label='A', step='pre') 23 ax.fill_between(df.start, df.e1, where=df.e1 < 0, color='tab:blue', ec='None', label='B', step='pre') ---> 24 ax.plot(df.start, z_assignment, color='tab:green', lw=1, label='z assignment') 26 # Add shaded areas for the nans 27 patches = [] File ~/miniconda3/envs/pymc/lib/python3.13/site-packages/matplotlib/axes/_axes.py:1777, in Axes.plot(self, scalex, scaley, data, *args, **kwargs) 1534 """ 1535 Plot y versus x as lines and/or markers. 1536 (...) 1774 (``'green'``) or hex strings (``'#008000'``). 1775 """ 1776 kwargs = cbook.normalize_kwargs(kwargs, mlines.Line2D) -> 1777 lines = [*self._get_lines(self, *args, data=data, **kwargs)] 1778 for line in lines: 1779 self.add_line(line) File ~/miniconda3/envs/pymc/lib/python3.13/site-packages/matplotlib/axes/_base.py:297, in _process_plot_var_args.__call__(self, axes, data, return_kwargs, *args, **kwargs) 295 this += args[0], 296 args = args[1:] --> 297 yield from self._plot_args( 298 axes, this, kwargs, ambiguous_fmt_datakey=ambiguous_fmt_datakey, 299 return_kwargs=return_kwargs 300 ) File ~/miniconda3/envs/pymc/lib/python3.13/site-packages/matplotlib/axes/_base.py:494, in _process_plot_var_args._plot_args(self, axes, tup, kwargs, return_kwargs, ambiguous_fmt_datakey) 491 axes.yaxis.update_units(y) 493 if x.shape[0] != y.shape[0]: --> 494 raise ValueError(f"x and y must have same first dimension, but " 495 f"have shapes {x.shape} and {y.shape}") 496 if x.ndim > 2 or y.ndim > 2: 497 raise ValueError(f"x and y can be no greater than 2D, but have " 498 f"shapes {x.shape} and {y.shape}") ValueError: x and y must have same first dimension, but have shapes (1460,) and (1438,)