import pandas as pd
import os
# Directory containing your .csv files
csv_dir = '../results/compartments/'
# Create a dictionary to store the DataFrames
dataframes = {}
# Iterate over all .csv files in the directory
for filename in os.listdir(csv_dir):
if filename.endswith('.csv'): # Check for .csv files
# Construct the full file path
filepath = os.path.join(csv_dir, filename)
# Load the CSV into a DataFrame
# Use the filename (without extension) as the dictionary key
key = filename.replace('_a_comp_coords_', '_')
key = os.path.splitext(key)[0]
dataframes[key] = pd.read_csv(filepath)
dataframes[key]['length'] = dataframes[key]['end'] - dataframes[key]['start']
# The `dataframes` dictionary now contains the DataFrames
dataframes.keys()
ech90 = pd.read_csv('../data/ech90_human_Mmul_10.csv')
ech90['length'] = ech90['end'] - ech90['start']E1 vs. ECH
Overlaying the A-Compartments with Extended Common Haplotypes
The genomic regions in question
In 03_compartments.ipynb we extracted the genomic intervals of A compartments on all cell types in all combinations of the following parameters:
- Cell type: fibroblast, spermatocyte, pachytene spermatocyte, round spermatid, sperm
- Chromosome: X
- E1 restriction: full-chromosome, chromosome arms, 10Mb windows
- Resolution: 100 kb, 500 kb
The following parameter was only changed for 100kb resolution:
- Smoothing: No smoothing, 5 bins (500kb)
Resulting in 45 .csv files. They are saved to ../results/compartments/.
Load the data
Time to unleash genominterv on the .csv files
Define a plotting function
# Kaspers plotting function
import pandas as pd
import matplotlib.pyplot as plt
import numpy as np
%config InlineBackend.figure_format = 'svg'
def plot_intervals(query=None, annot=None, **kwargs):
tups = []
if query is not None:
tups.append(('query', query))
if annot is not None:
tups.append(('annot', annot))
tups.extend(kwargs.items())
tups = reversed(tups)
df_list = []
labels = []
for label, df in tups:
labels.append(label)
df['label'] = label
df_list.append(df)
bigdf = pd.concat(df_list)
bigdf['chrom'] = pd.Categorical(bigdf['chrom'], bigdf['chrom'].unique())
bigdf['label'] = pd.Categorical(bigdf['label'], bigdf['label'].unique())
gr = bigdf.groupby('chrom', observed=False)
fig, axes = plt.subplots(gr.ngroups, 1, figsize=(8, 1.5*gr.ngroups),
sharey=True
# sharex=True
)
if type(axes) is not np.ndarray:
# in case there is only one axis so it not returned as a list
axes = np.array([axes])
# with plt.style.context(('default')):
for i, chrom in enumerate(gr.groups):
_df = gr.get_group(chrom)
_gr = _df.groupby('label', observed=False)
for y, label in enumerate(_gr.groups):
try:
df = _gr.get_group(label)
except KeyError:
continue
y = np.repeat(y, df.index.size)
axes[i].hlines(y, df.start.tolist(), df.end.tolist(), alpha=0.5, lw=5, colors=f'C{y[0]}')
delta = len(labels)/10
axes[i].vlines(df.start.tolist(), y-delta, y+delta, alpha=0.5, lw=2, colors=f'C{y[0]}')
axes[i].vlines(df.end.tolist(), y-delta, y+delta, alpha=0.5, lw=2, colors=f'C{y[0]}')
axes[i].spines['top'].set_visible(False)
axes[i].spines['left'].set_visible(False)
axes[i].spines['right'].set_visible(False)
axes[i].set_yticks(list(range(len(labels))), labels)
axes[i].tick_params(axis='y', which='both', left=False)
axes[i].set_ylim(-1, len(labels)-0.7)
# axes[i].set_xlim(df.start.min()-delta, df.end.max()+delta)
if i != gr.ngroups-1:
axes[i].tick_params(axis='x', which='both', bottom=False)
axes[i].set_title(chrom, loc='left', fontsize=10)
plt.tight_layout()# My plotting function
import matplotlib.pyplot as plt
import numpy as np
import pandas as pd
from matplotlib.patches import Rectangle
from mpl_toolkits.axes_grid1 import make_axes_locatable
from genominterv import interval_intersect
%config InlineBackend.figure_format = 'svg'
def plot_regions(query=None, annot=None, intersect=None):
chrom = annot['chrom'].unique()[0]
chromsize = pd.read_csv('../data/rheMac10.filtered.chrom.sizes', sep='\t', header=None, names=['chrom', 'size'])
chromsize = chromsize[chromsize['chrom'] == chrom]['size'].values[0]
# Define the plot
height = 1 + (1 if query is not None else 0) + (1 if intersect is not None else 0)
height = height * 0.75
f, ax = plt.subplots(figsize=(10, height), sharex=True)
ax.spines[:].set_visible(False)
# Plot the annot
# Iterate over each interval in the DataFrame
for start, end in zip(annot['start'], annot['end']):
rect = Rectangle((start, 0.1), width=end-start, height=0.9, color='tab:red', linewidth=0, alpha=0.6)
ax.add_patch(rect)
ax.spines['bottom'].set_visible(True)
lbl = annot['label'].unique()[0] if 'label' in annot.columns else 'A-Comp'
ax.set_ylabel(lbl, rotation=0, fontsize=10, labelpad=30)
divider = make_axes_locatable(ax)
if query is not None:
qax = divider.append_axes("top", size="100%", pad=0.2, sharex=ax)
qax.xaxis.set_visible(False)
# Plot the query
for start, end in zip(query['start'], query['end']):
rect = Rectangle((start, 0.1), width=end-start, height=0.9, color='tab:blue', linewidth=0, alpha=0.6)
qax.add_patch(rect)
qax.spines[:].set_visible(False)
qax.set_yticks([])
qax.set_title(chrom, loc='left', fontsize=10)
qax.set_ylabel('ECH90', rotation=0, fontsize=10, labelpad=30)
if intersect is not None:
iax = divider.append_axes("bottom", size="100%", pad=0.2, sharex=ax)
# Invisible x-axis for 'annot' (intersect ie below)
ax.xaxis.set_visible(False)
# Plot the intersect
for start, end in zip(intersect['start'], intersect['end']):
rect = Rectangle((start, 0.1), width=end-start, height=0.9, color='tab:green', linewidth=0, alpha=0.6)
iax.add_patch(rect)
iax.spines[:].set_visible(False)
iax.set_yticks([])
ax.spines['bottom'].set_visible(False)
iax.spines['bottom'].set_visible(True)
iax.set_ylabel('Intersect', rotation=0, fontsize=10, labelpad=30)
ax.set_yticks([])
ax.set_xlim(0, chromsize)
ticks = np.linspace(0, chromsize, num=5)
ax.set_xticks(ticks)
ax.set_xticklabels([f'{int(t/1e6)} Mbp' for t in ticks])
plt.tight_layout()
return f, ax
Test with a subsample of the data
annot = dataframes['round_spermatid_100kb_arms']
query = ech90
intersect = interval_intersect(annot, query)
plot_intervals(query, annot, intersection=intersect)
plot_regions(query, annot, intersect)from genominterv import proximity_test, interval_collapse, interval_diff, interval_intersect, jaccard_stat
annot = dataframes['round_spermatid_100kb_arms']
query = ech90
#plot_intervals(query=query, annot=annot)
for key,annot in dataframes.items():
# Filter out subset
if ('round_spermatid_100') in key and not 'full' in key:
# Plot the intervals
intersection = interval_intersect(query, annot)
plot_intervals(query=query, annot=annot, intersection=intersection)
plt.title(key)
# Do a proximity test
print(f"Tests for {key}")
annot_collapsed = interval_collapse(annot)
non_ovl_query = interval_diff(query, annot_collapsed)
print("Proximity:", proximity_test(non_ovl_query, annot_collapsed))
print("Jaccard:", jaccard_stat(query, annot))
print()
Tests for round_spermatid_100kb_arms_smoothed
Proximity: TestResult(statistic=0.20566666666666641, pvalue=0.105)
Jaccard: 0.03319511172796144
Tests for round_spermatid_100kb_arms
Proximity: TestResult(statistic=0.49242857142857144, pvalue=0.0004)
Jaccard: 0.03916232332293147
Tests for round_spermatid_100kb_10Mb
Proximity: TestResult(statistic=0.3223076923076922, pvalue=0.0209)
Jaccard: 0.04512778341139746
Tests for round_spermatid_100kb_10Mb_smoothed
Proximity: TestResult(statistic=0.4658333333333337, pvalue=0.0017)
Jaccard: 0.04494391747197651
Bootstrap to get a p-value
from genominterv import bootstrap
annot = dataframes['round_spermatid_100kb_arms']
chromsizes = (
pd.read_csv('../data/rheMac10.filtered.chrom.sizes',
sep='\t',
index_col='chrom',
header=None,
names=['chrom','size'])
.to_dict()['size']
)
#display(chromsizes)
@bootstrap(chromsizes, samples=1000)
def jaccard_bootstrap(query, annot):
return jaccard_stat(query, annot)
jacccard_stat, p_value = jaccard_bootstrap(query, annot)jacccard_stat, p_value(0.03916232332293147, 0.31)Partition the A-compartments into regions around the edges
df = dataframes['round_spermatid_100kb_arms']
start_edge = pd.DataFrame({
'chrom': df['chrom'],
'start': df['start']-1*df['resolution'],
'end': df['start']+1*df['resolution'],
'resolution': df['resolution'],
'label': 'start_edge'
})
end_edge = pd.DataFrame({
'chrom': df['chrom'],
'start': df['end']-1*df['resolution'],
'end': df['end']+1*df['resolution'],
'resolution': df['resolution'],
'label': 'end_edge'
})
#df
test_df = pd.concat([start_edge, end_edge]).sort_values(['chrom', 'start', 'end'])
interval_collapse(test_df)| start | end | chrom | |
|---|---|---|---|
| 0 | 800000 | 1000000 | chrX |
| 1 | 2500000 | 2700000 | chrX |
| 2 | 3100000 | 3300000 | chrX |
| 3 | 3400000 | 3600000 | chrX |
| 4 | 6500000 | 6800000 | chrX |
| ... | ... | ... | ... |
| 86 | 147000000 | 147200000 | chrX |
| 87 | 148800000 | 149400000 | chrX |
| 88 | 150800000 | 151000000 | chrX |
| 89 | 152200000 | 152400000 | chrX |
| 90 | 153200000 | 153400000 | chrX |
91 rows × 3 columns
import os
for key, df in dataframes.items():
outdir = '../results/edges'
edge_csv_name = os.path.join(outdir,f'{key+'_edges.csv'}')
if not os.path.exists(edge_csv_name):
res = df['resolution'].unique()[0]
start_edge = pd.DataFrame({
'chrom': df['chrom'],
'start': df['start']-1*df['resolution'],
'end': df['start']+1*df['resolution'],
'resolution': df['resolution'],
'label': 'start_edge'
})
end_edge = pd.DataFrame({
'chrom': df['chrom'],
'start': df['end']-1*df['resolution'],
'end': df['end']+1*df['resolution'],
'resolution': df['resolution'],
'label': 'end_edge'
})
if not os.path.exists(outdir):
os.makedirs(outdir)
tmp = pd.concat([start_edge, end_edge]).sort_values(['chrom', 'start', 'end'])
interval_collapse(tmp).assign(resolution=res).to_csv(edge_csv_name, index=False)Import edges
import pandas as pd
import os
# Directory containing your .csv files
csv_dir = '../results/edges/'
# Create a dictionary to store the DataFrames
edges = {}
# Iterate over all .csv files in the directory
for filename in os.listdir(csv_dir):
if filename.endswith('.csv'): # Check for .csv files
# Construct the full file path
filepath = os.path.join(csv_dir, filename)
# Load the CSV into a DataFrame
# Use the filename (without extension) as the dictionary key
key = filename.replace('_edges_', '')
key = os.path.splitext(key)[0]
edges[key] = pd.read_csv(filepath)
edges[key]['length'] = edges[key]['end'] - edges[key]['start']
# The `edges` dictionary now contains the DataFrames
print(edges.keys())
print(edges['fibroblast_100kb_10Mb_edges'].columns)
#ech90 = pd.read_csv('../data/ech90_human_Mmul_10.csv')dict_keys(['sperm_100kb_arms_smoothed_edges', 'spermatogonia_500kb_full_edges', 'pachytene_spermatocyte_100kb_10Mb_smoothed_edges', 'spermatogonia_100kb_arms_edges', 'fibroblast_500kb_full_edges', 'round_spermatid_500kb_10Mb_edges', 'fibroblast_100kb_arms_edges', 'spermatogonia_100kb_full_smoothed_edges', 'round_spermatid_100kb_full_smoothed_edges', 'round_spermatid_100kb_full_edges', 'fibroblast_100kb_10Mb_edges', 'round_spermatid_500kb_arms_edges', 'fibroblast_100kb_full_smoothed_edges', 'spermatogonia_100kb_10Mb_edges', 'sperm_500kb_arms_edges', 'spermatogonia_100kb_10Mb_smoothed_edges', 'sperm_100kb_full_edges', 'pachytene_spermatocyte_500kb_10Mb_edges', 'pachytene_spermatocyte_100kb_full_smoothed_edges', 'pachytene_spermatocyte_100kb_full_edges', 'pachytene_spermatocyte_500kb_arms_edges', 'fibroblast_100kb_10Mb_smoothed_edges', 'sperm_500kb_10Mb_edges', 'round_spermatid_100kb_10Mb_smoothed_edges', 'spermatogonia_500kb_arms_edges', 'spermatogonia_100kb_full_edges', 'spermatogonia_100kb_arms_smoothed_edges', 'fibroblast_500kb_arms_edges', 'round_spermatid_100kb_10Mb_edges', 'fibroblast_100kb_full_edges', 'sperm_100kb_full_smoothed_edges', 'fibroblast_100kb_arms_smoothed_edges', 'round_spermatid_100kb_arms_edges', 'fibroblast_500kb_10Mb_edges', 'round_spermatid_500kb_full_edges', 'round_spermatid_100kb_arms_smoothed_edges', 'spermatogonia_500kb_10Mb_edges', 'sperm_500kb_full_edges', 'sperm_100kb_10Mb_smoothed_edges', 'sperm_100kb_arms_edges', 'pachytene_spermatocyte_100kb_arms_smoothed_edges', 'pachytene_spermatocyte_100kb_10Mb_edges', 'pachytene_spermatocyte_100kb_arms_edges', 'pachytene_spermatocyte_500kb_full_edges', 'sperm_100kb_10Mb_edges'])
Index(['chrom', 'start', 'end', 'resolution', 'label', 'length'], dtype='object')from genominterv import interval_intersect
sample = 'round_spermatid_100kb_arms'
full_df = dataframes[sample]
full_intersect = interval_intersect(full_df, ech90).assign(length=lambda x: x['end'] - x['start'])
edge_df = edges[f'{sample}_edges']
# Plot full
plot_regions(ech90, full_df, full_intersect)
plt.suptitle('All edges')Text(0.5, 0.98, 'All edges')### Some stats about the data and intersections ###
# Determine the proportion of total regions in ECH90 that lies on compartment edges
print("Proportion of ECH90 on edges (#count)")
# Proportion of ECH90 on full regions
print(f'\t{full_intersect.shape[0] / ech90.shape[0]}')
print("\nProportion of ECH90 on edges (#bpairs)")
# Proportion of ECH90 on full regions
print(f'\t{full_intersect['length'].sum() / ech90['length'].sum()}')
# What is the total length of ech90 regions
print("\nTotal length of:")
print(f'\tECH90: {(ech90['end'] - ech90['start']).sum()} bp')
print(f'\tEdges: {(edge_df["end"] - edge_df["start"]).sum()} bp')Proportion of ECH90 on edges (#count)
0.631578947368421
Proportion of ECH90 on edges (#bpairs)
0.3932398045966063
Total length of:
ECH90: 5511675 bp
Edges: 28800000 bpf, ax = plot_regions(edge_df, ech90, full_intersect)
for ax in list(f.axes):
if ax.get_ylabel() == 'ECH90':
ax.set_ylabel('edge_df', rotation=0, fontsize=10, labelpad=30)
if ax.get_ylabel() == "query":
ax.set_ylabel('ECH90', rotation=0, fontsize=10, labelpad=30)
if ax.get_ylabel() == 'Intersect':
#ax.set_ylabel('end_edge \nintersect', rotation=0, fontsize=10, labelpad=30)
pass
Do testing of the edges
%%capture
# Define what we are testing
print("""
Goal: To test whether ECH90 regions are enriched in compartment edges
Query: ECH90
Annotation: Start and end edges of compartments
Hypothesis:
ECH90 regions are enriched in compartment edges
Null hypothesis:
ECH90 regions are not enriched in compartment edges
Tests:
* Proximity test:
tests whether the query regions are closer to
the annotation regions than expected by chance.
NB regions can not overlap, so we need to collapse the annotation regions
* Jaccard index:
tests the similarity between the query and annotation regions,
where a value of 1 indicates perfect overlap
""")### Proximity test ###
from genominterv import proximity_test, interval_collapse, interval_diff, interval_intersect
# Define the query and annotation
query = ech90
annot = full_intersect
# Calculate the non-overlapping query regions
non_ovl_query_full = interval_diff(query, annot)
# Perform the proximity test
proximity_full = proximity_test(non_ovl_query_full, annot, two_sided=False)
print("Proximity test results: All edges")
print(f"\tstatistic: {proximity_full.statistic}, \n\tp-value: {proximity_full.pvalue}")Proximity test results: All edges
statistic: 0.6821666666666665,
p-value: 0.0### Jaccard index ###
from genominterv import jaccard_stat, bootstrap
chromsizes = (pd.read_csv(
'../data/rheMac10.filtered.chrom.sizes',
sep='\t',
index_col='chrom',
header=None,
names=['chrom','size'])
.to_dict()['size']
)
# # Calculate the Jaccard index
# jaccard_start = jaccard_stat(query, annot_start)
# jaccard_end = jaccard_stat(query, annot_end)
# jaccard_concat = jaccard_stat(query, annot_concat)
# print("\nJaccard index results")
# print(f"Start edge: {jaccard_start}")
# print(f"End edge: {jaccard_end}")
# print(f"Concat edge: {jaccard_concat}")
# Test with bootstrap decorator
@bootstrap(chromsizes, samples=1000)
def jaccard_bootstrap(query, annot):
return jaccard_stat(query, annot)
jaccard_stat_full, p_value_full = jaccard_bootstrap(query,annot)
print(f"Jaccard index: {jaccard_stat_full}, p-value: {p_value_full}")Jaccard index: 0.3932398045966063, p-value: 0.0p-value is zero smaller than 0.001. Increase nr samples to get actual p-value.Check the length of the intervals
Bioframe genomic intervals support
import bioframe Geneinfo
How does the edges align with genes?
This first plot is just to figure out how to plot with gene_plot.
import geneinfo as gi
from matplotlib.patches import Rectangle
from matplotlib.collections import PatchCollection
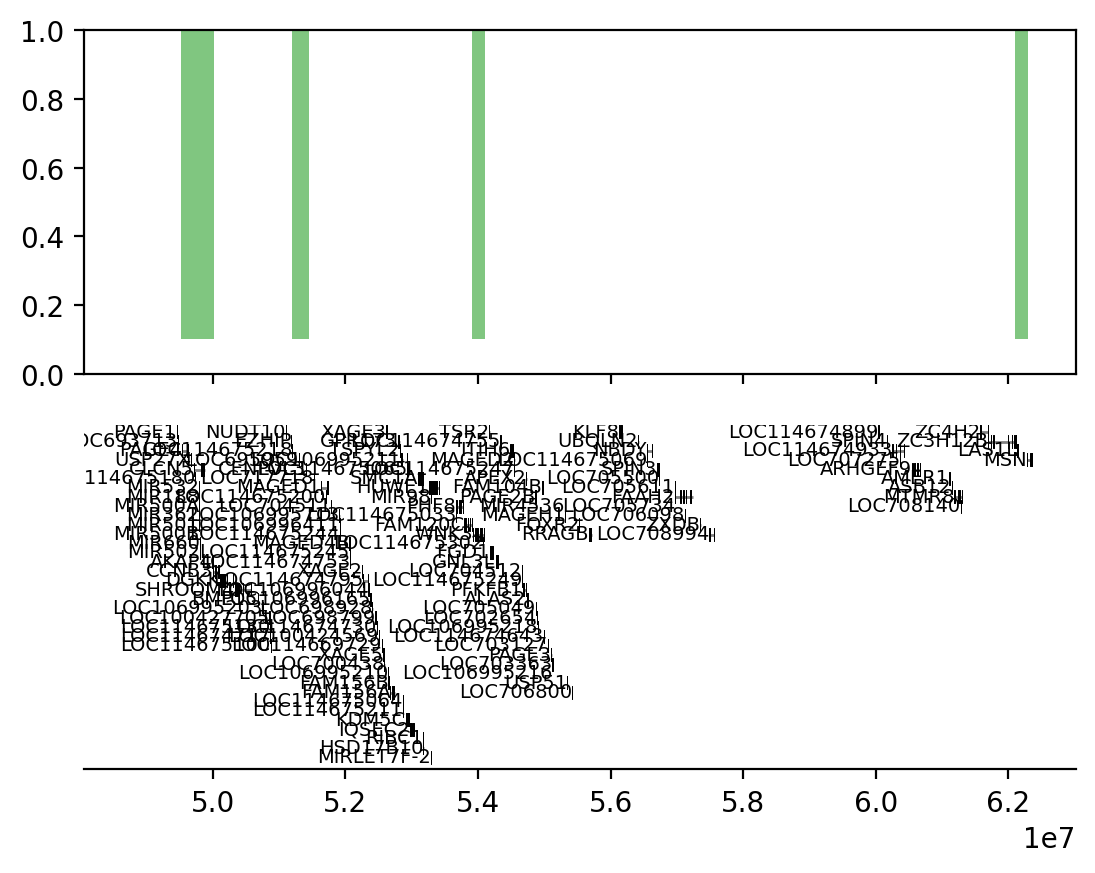
# Use the proximity test results to plot the ECH90 regions and the compartment edges
start = full_intersect['start'][2]
end = full_intersect['end'][5]
rectangles = [Rectangle((start, 0.1), width=end-start, height=0.9, color='tab:green', linewidth=0, alpha=0.6) for start, end in zip(full_intersect['start'][2:6], full_intersect['end'][2:6])]
pc = PatchCollection(rectangles, match_original=True)
ax = gi.gene_plot('chrX', start-100_000, end+100_000, assembly='rheMac10')
ax.add_collection(pc)Get the geneinfo for all intersections between edges and ECH90
And write to a csv file. If the file exists, read it with pandas.
# Use get_genes_region
import os.path as op
import geneinfo as gi
import pandas as pd
genes_file = '../results/edge_genes/rs_edges_100kb_genes.csv'
if not op.exists(genes_file):
genes = pd.concat(
full_intersect.apply(
lambda x: gi.get_genes_region_dataframe('chrX', x['start'], x['end'], assembly='rheMac10'),
axis =1
).to_list(),
ignore_index=True
)
genes.to_csv(genes_file, index=False)
else:
genes = pd.read_csv(genes_file)genes_list = genes['name'].unique().tolist()
genes_list['SH3KBP1',
'MIR7206',
'LANCL3',
'XK',
'CYBB',
'LOC696657',
'DYNLT3',
'PAGE4',
'USP27X',
'CLCN5',
'LOC114675180',
'MIR532',
'MIR188',
'MIR500A',
'MIR362',
'MIR501',
'MIR500B',
'MIR660',
'MIR502',
'AKAP4',
'CCNB3',
'LOC114675218',
'LOC695959',
'CENPVL3',
'FAM120C',
'WNK3',
'LOC114675302',
'ZC3H12B',
'LAS1L',
'MSN',
'ATRX',
'MAGT1',
'LOC114675151',
'COX7B',
'ATP7A',
'ALG13',
'RAP2C',
'DKC1',
'LOC114675231',
'MPP1',
'SMIM9',
'F8']import geneinfo as gi
from matplotlib.patches import Rectangle
from matplotlib.collections import PatchCollection
# Use the proximity test results to plot the ECH90 regions and the compartment edges
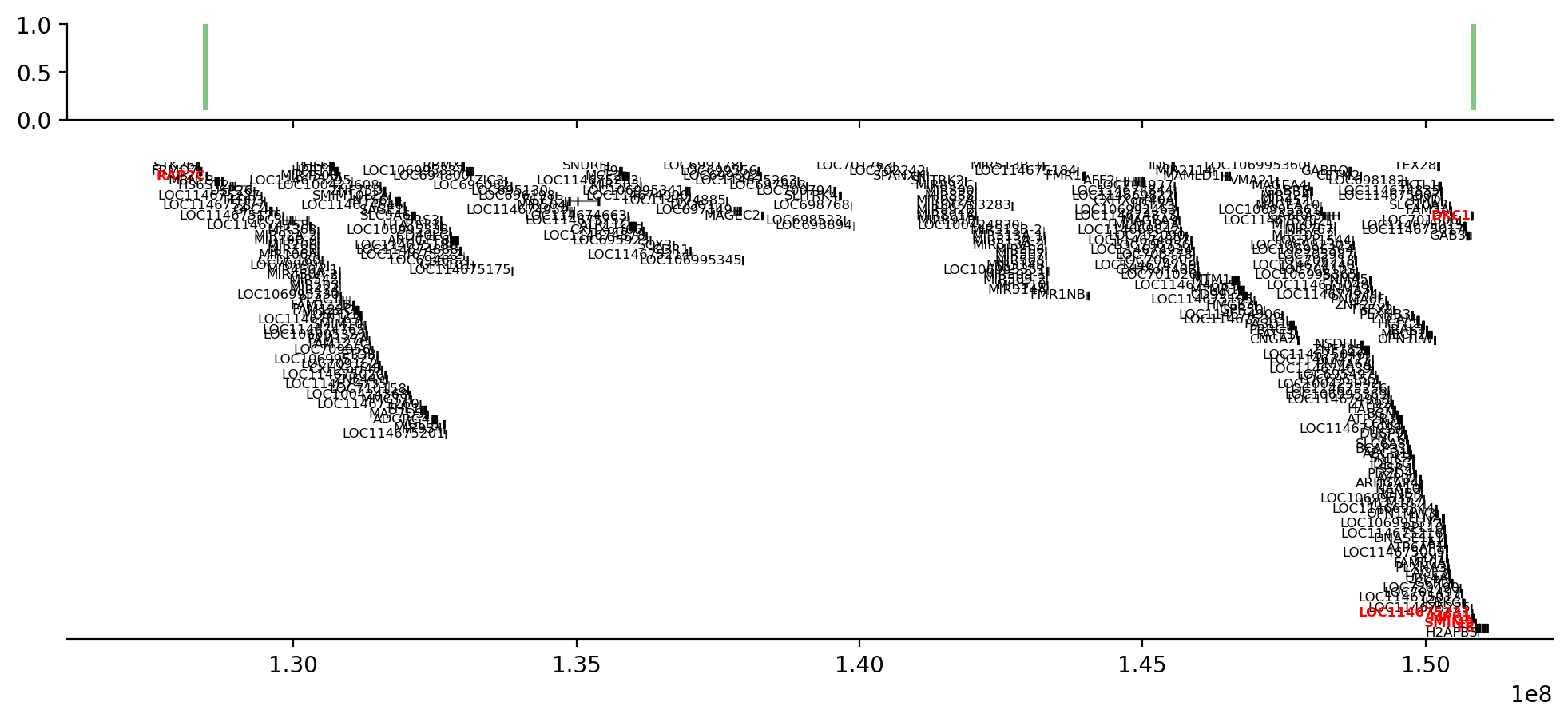
start_idx = 10
end_idx = 11
start = full_intersect['start'][start_idx]
end = full_intersect['end'][end_idx]
rectangles = [Rectangle(
(start, 0.1), width=end-start, height=0.9, color='tab:green', linewidth=0, alpha=0.6) for start, end in zip(full_intersect['start'][start_idx:end_idx+1], full_intersect['end'][start_idx:end_idx+1])]
pc = PatchCollection(rectangles, match_original=True)
ax = gi.gene_plot('chrX', start-100_000, end+100_000, assembly='rheMac10',
highlight=genes_list,
despine=True,
figsize=(12, 5),
aspect=5,
)
ax.add_collection(pc)What can I do with the list of genes on the edges?
GO enrichment?
mmul_x_genes = gi.get_genes_region_dataframe('chrX', 0, 155_000_000, assembly='rheMac10')mmul_x_genelist = mmul_x_genes['name'].unique().tolist()gene_list = genes['name'].unique().tolist()
taxid = 9544
gi.email('sojernj@gmail.com')
#gi.go_annotation_table(taxid=taxid)
#gi.show_go_evidence_codes()
go_terms = gi.get_go_terms_for_genes(gene_list, taxid=taxid)len(go_terms)
#gene_list[:5]98results = gi.go_enrichment(
gene_list[:5],
# Use human as a start
alpha=0.05,
terms=go_terms
)geneinfo_cache/go-basic.obo: fmt(1.2) rel(2024-10-27) 44,017 Terms; optional_attrs(def relationship)Could not map gene symbol "MIR7206" to ncbi id