29 GWAS candidates
The purpose of this exercise is to expose you to the different kinds of information that are stored in databases relevant to bioinformatics. It takes experience and skill to navigate the user interface of these databases. Here, you will see a few important ones, but there are many others. I have put a list at the end of this exercise with some additional relevant ones on Brightspace. Browse them at your heart’s content.
Genome-wide association studies (GWAS) are a powerful tool in genetics and genomics research to identify genetic variants associated with diseases or traits. When a significant SNP is identified in a GWAS, it is a genomic signpost tagging an associated genomic region. Still, it only partially reveals the genes causing the disease. The genomic region identified likely contains several genes or genomic features. Investigating candidate disease genes near a GWAS SNP involves: Exploring the genomic region around the SNP. Studying nearby genes and regulatory elements. Evaluating their potential roles in the disease or trait of interest. A standard GWAS only includes a select subset of the SNPs in the genome. So, the most significant included SNP variant is rarely responsible for the disease. Still, it is so close to the causal one that individuals with the causal SNP variant carry it. The closer two SNPs are along the genome, the more likely they appear together like this. The further away from each other, the more likely it is that genetic recombination has removed this correlation.
Type 1 diabetes, often referred to as juvenile diabetes or insulin-dependent diabetes, is a chronic autoimmune condition that affects how the body regulates blood sugar (glucose). Unlike type 2 diabetes, which is commonly associated with lifestyle factors such as obesity and physical inactivity, type 1 diabetes is not preventable and typically develops early in life, often during childhood or adolescence. In type 1 diabetes, the immune system, usually responsible for defending the body against harmful invaders like bacteria and viruses, becomes misguided. It mistakenly identifies the insulin-producing beta cells within the pancreas as foreign threats. This misrecognition triggers an autoimmune response, leading immune cells to launch an attack on these vital beta cells. The pancreas, an organ located behind the stomach, is a key player in regulating blood sugar levels. It consists of clusters of cells called the Islets of Langerhans, which house the insulin-producing beta cells. When the immune system attacks these beta cells, it results in their destruction or severe impairment. This process is thought to involve various immune cells, such as T-cells and antibodies, which play pivotal roles in autoimmune reactions. Due to this autoimmune attack, the pancreas gradually loses its ability to produce insulin, a hormone with crucial responsibilities in maintaining proper blood sugar balance. Insulin acts as a molecular key that unlocks the doors of cells throughout the body, allowing glucose to enter and be used as an energy source. Think of insulin as a bridge that facilitates the movement of glucose from the bloodstream into cells. Research into type 1 diabetes is ongoing, and scientists are exploring potential cures and better management strategies. However, until a cure is found, individuals with type 1 diabetes continue to lead fulfilling lives by effectively managing their condition through insulin therapy, a balanced diet, regular exercise, and closely monitoring their blood sugar levels.
Browsing SNPs from a GWAS
Start by examining a Manhattan plot or list of significant SNPs obtained from a GWAS study. We are going to select one of these SNPs and do further analysis. While doing the analysis, note the SNP identifier (rsID) and its significance level (p-value). Go to locuszoom.org and press the blue button shown on Figure 29.1:
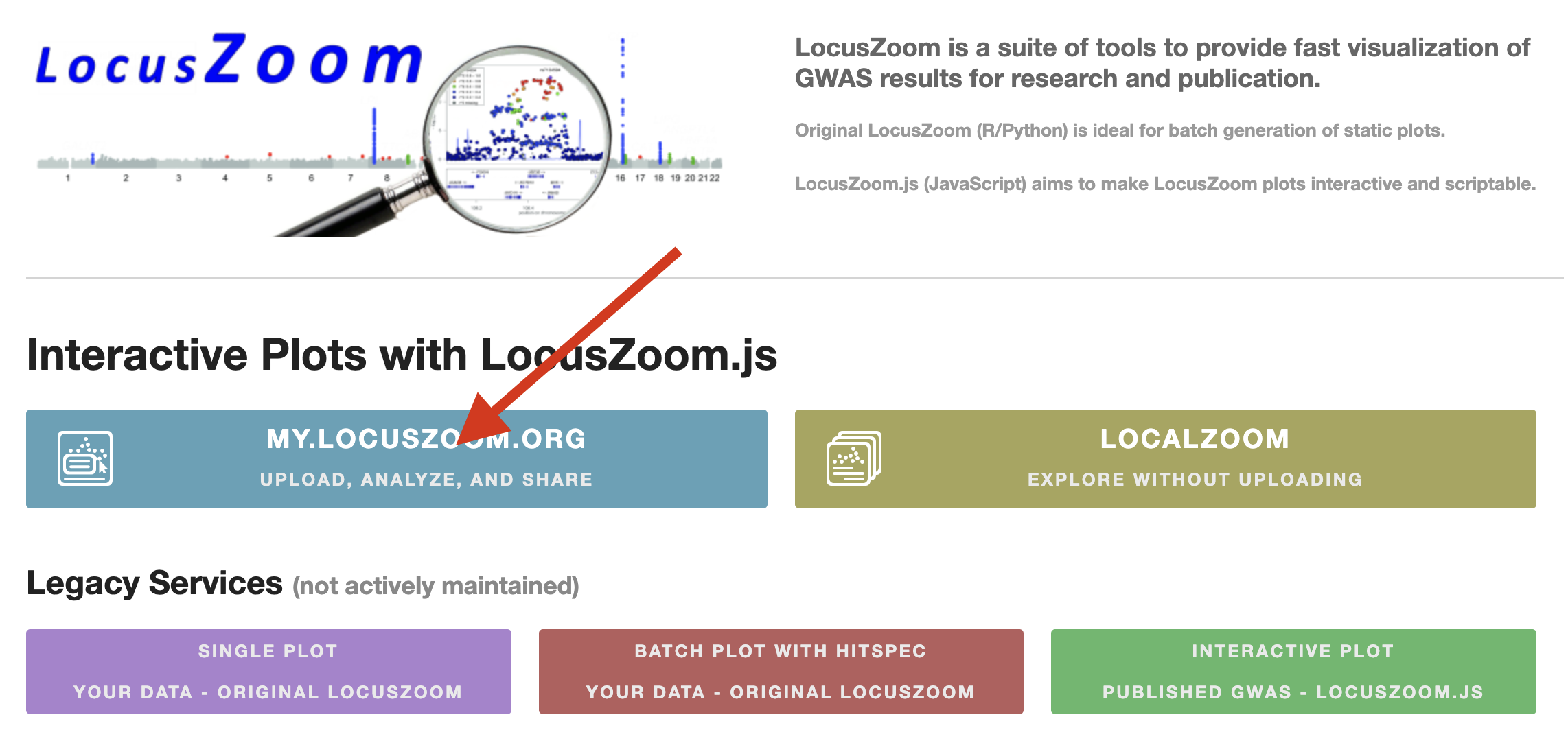
Now, you should be able to search for public GWAS studies. Copy/paste “Rare Genetic Variants of Large Effect Influence Risk of Type 1 Diabetes” into the search bar. Only one result should show up. Press it. Now, you are ready to examine the Manhattan plot. You can see a lot of significant SNPs in the Manhattan plot, and you are welcome to examine them, but the one we are interested in for this exercise is the one located close to RSBN1. Click it, and it will zoom in. The SNP has an rsID of rs6679677, and its position is 113.761.186 on chromosome 1. It’s a C to A mutation. Save this information because we will use it for the rest of the exercise.
Exploring a genomic region
In this section, we are going to explore our SNP and its surrounding genomic regions to identify our candidate gene/genes for diabetes type 1. Go to the UCSC Genome Browser. In the “Human Assembly” drop-down, make sure it says “GRh38/hg38”. That way, you access the genome assembly also used in the GWAS study. If you choose another one, the SNP and gene coordinates will be different. Now paste your SNP identifier rs6679677 into the search field where it says “Position/Search Term” and click GO. You will get a list of search results. Clicking the link in the top one (if it says rs6679677) should take you to a page centered on this SNP looking something like the one shown in Figure 29.2:
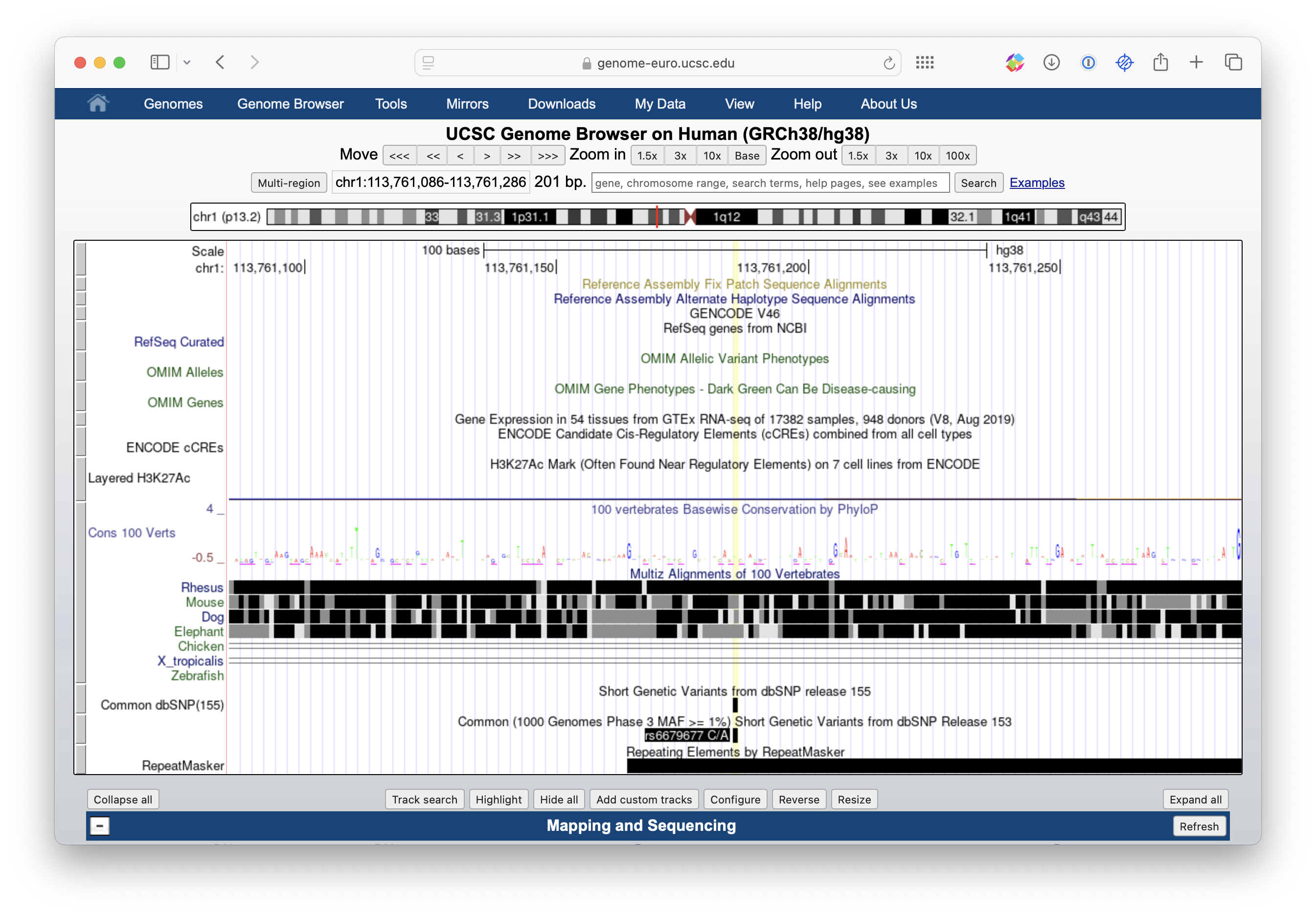
Now, you see the SNP in a window. You can play around with the zoom function until you start seeing a couple of other SNP IDs. Now we want to look at surrounding genes, so scroll down to the “Genes and Gene predictions” Section. All the options you see here are called “tracks”, and they all contain different information about the genome. If you want to look at the annotated gene, select the “GENCODE V48” select “pack” and press refresh on the right side of the section. Play around with the different settings (pack, dense, squish, all). They all give you a different amount of information. Select the one that you are most comfortable with when exploring. You might have to change it for some types of questions. “Pack” is a good place to start.
If there is too much going on for your liking, you can hide certain tracks at the bottom of the page. Here is a link that will lead you to the browser, with all but the gene and SNP tracks hidden: [UCSC Genome Browser with hidden tracks] (https://genome-euro.ucsc.edu/cgi-bin/hgTracks?db=hg38&lastVirtModeType=default&lastVirtModeExtraState=&virtModeType=default&virtMode=0&nonVirtPosition=&hgsid=407916288_fkUQBcFRTUFVP1XZP2VxzusTgHLA)
Exercise 29-1
Play around with the zoom function. To the left of where you inserted the SNP ID is the size of the window you are looking at. The more you zoom out, the more genes you will be able to see.
Exercise 29-2
Identify nearby genes, including their names, locations, and orientations. Remember that linkage means that the causal gene may not be the one closest to the GWAS SNP. Are there any genes located close to the SNP of interest? If yes. Note them down for further analysis.
Exercise 29-3
What is the genomic context (e.g., intronic, intergenic) of the SNP?
Exercise 29-4
You see the same gene multiple times in the gene track. Why do you think that is?
Exercise 29-5
Try to select other tracks such as promoters (EPDnew Promoters) and regulatory elements (ENCODE Regulation). See if there are known regulatory elements, such as enhancers or promoters, in the vicinity of the SNP.
Exercise 29-6
Are there any annotated regulatory elements near the SNP that might influence gene expression? Try to press the regulatory track. What kind of information does this track represent? (Hint: Read the description of the track).
Gathering information about genes
Now, we are ready to look at the genes we are interested in relation to our SNP. For this, we will use 3 well-known databases. The goal is to identify which genes are the potential cause of type 1 diabetes. Take your genes to each of the websites and use the information they give you to see which genes can be related to diabetes.
It’s important to understand what kind of information these sites provide and their intentions. So, let us do some light exploration.
Exercise 29-7
Google “NCBI”. What is it, and who administers it? Do the same with ENSEMBL. What’s its function, and who administers it?
Exercise 29-8
On the NCBI Gene website, the drop-down menu shows “gene”, but other databases on NCBI exist. Get yourself an overview. Do you recognize other databases? Some you don’t know?
Exercise 29-9
When a gene is found in the “About this gene” section at ENSEMBL, try to press “phenotypes” if there are any. Do you see diabetes for any of the genes?
Exercise 29-10
The Genecards resource is different from the two other databases. Can you list two major differences?
Exercise 29-11
By now, you should have a feeling for the databases and their goals. NCBI and ENSEMBL store the same sequence information, so it is mostly about which site presents data in the most useful way for the task at hand. Which one is your favorite one and why? Which of the genes are related to Type 1 Diabetes? If you can’t find a related gene, go back to “Accessing the UCSC Genome Browser and make initial exploration” and look at your tracks again. You may have missed it.
Exercise 29-12
With the gene you now have chosen, answer the following questions by using the 3 above databases.
- How long is the entire of the gene (in bp or kb)?
- How many transcripts of the gene are in NCBIs “Transcript table”?
- Does Ensembl and NCBI show the same number of transcripts?
- In the graphical representation of a gene, how are exons and introns depicted at ENSEMBL and NCBI?
- At ENSEMBL, try to click on the UniprotKN identifier. This takes you to a page in the “UniProt” database. Can you retrive the amino acid sequence? - Do that.
Exploring tissue expression
In this step, we are interested in investigating our gene’s expression. Or multiple genes if we have yet to choose a specific gene. Expression analysis will help us understand the genes better. Go to the GTEx Portal (https://www.gtexportal.org/home/) and search for the gene/genes associated with your SNP to see their expression profiles across tissues. Explore tissue-specific gene expression data for the identified genes. In which tissues is the gene(s) expressed, and is there any tissue where its expression is notably higher? You may know “EBV-transformed lymphocytes” as “lymphocytes” cells. They are important to the immune system.
Exercise 29-13
In which tissues is the gene(s) typically expressed, and are there variations in expression levels across different tissues?
Exercise 29-14
Do the expression profiles make sense with the information you got from the other databases? Do you expect to see certain tissues with a high expression? Other with a low? Is it relevant to the disease we are investigating?
Exercise 29-15
What can you infer about the potential functional relevance of the gene(s) in the context of the disease or trait?
Hypotheses and Discussion
Based on the information gathered from UCSC, various gene databases, and GTEx, propose hypotheses about the potential role of the identified gene(s) in the disease or trait associated with the SNP. Think about the importance of tissue-specific gene expression in understanding disease mechanisms. Are there any other genes or regulatory elements near the SNP that could also be relevant to the disease or trait?
Exercise 29-16
Based on the genomic and expression data, what are your hypotheses regarding the role of the identified gene(s) in the disease or trait?
Exercise 29-17
How might tissue-specific gene expression patterns provide insights into the disease mechanism?
Exercise 29-18
Are there potential challenges or limitations in establishing causality between the gene(s) and the disease?
Exercise 29-19
Summarize your findings and discuss the significance of investigating candidate disease genes near a GWAS SNP. Reflect on the challenges and limitations of this approach in pinpointing causal genes for complex diseases.
Exercise 29-20
What did you learn from this exercise about the importance of exploring the genomic context and tissue-specific gene expression in GWAS follow-up?
Exercise 29-21
How might these findings inform future research or therapeutic approaches for the disease or trait?